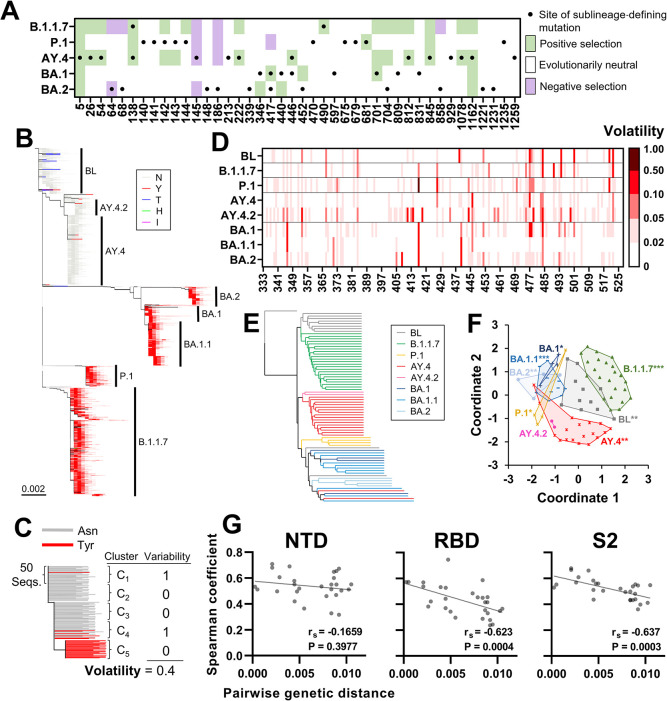
Fig 1. The mutational space of spike is specific for SARS-CoV-2 lineage.
(A) Black dots indicate sites of mutation that define VOC sublineages and emerged until April 8th 2022. Cells are shaded by the inferred selective pressures applied on the sites in the baseline of each VOC calculated using the SLAC method. (B) Phylogenetic tree based on 40,350 unique spike sequences from the indicated lineages. The SARS-CoV-2 baseline (BL) group is composed of sequences within 0.0015 nucleotide substitutions per site from the SARS-CoV-2 ancestral strain. Branches are colored by the residue at position 501. (C) Schematic of the approach to calculate volatility for each position of spike. (D) Volatility at RBD positions in the indicated VOCs or the BL group. (E) Lineages were partitioned into groups of 500 sequences and the absence or presence of volatility at all spike positions in each group was determined. All groups were thus assigned 1273-bit strings that describes the volatility profile of spike. Strings were compared using the UPGMA clustering method. (F) Relationships between the 1273-bit strings. Data points represent the strings of all 500-sequence clusters, which are labeled by lineage. To visualize these relationships, the distance matrix between all vectors was used as input for multidimensional scaling. Lineage-specificity of the profiles was determined by a permutation test. P-values: *, P<0.05; **, P<0.005; ***, P<0.0005. (G) Volatility was calculated for each lineage at all positions of the NTD (20–286), RBD (333–527) and S2 (686–1213). The Spearman correlation coefficient between volatility values in any two lineages was determined. Coefficients are compared with the mean nucleotide distance that separates any two lineages. rS, Spearman coefficient. P-values, two-tailed test.