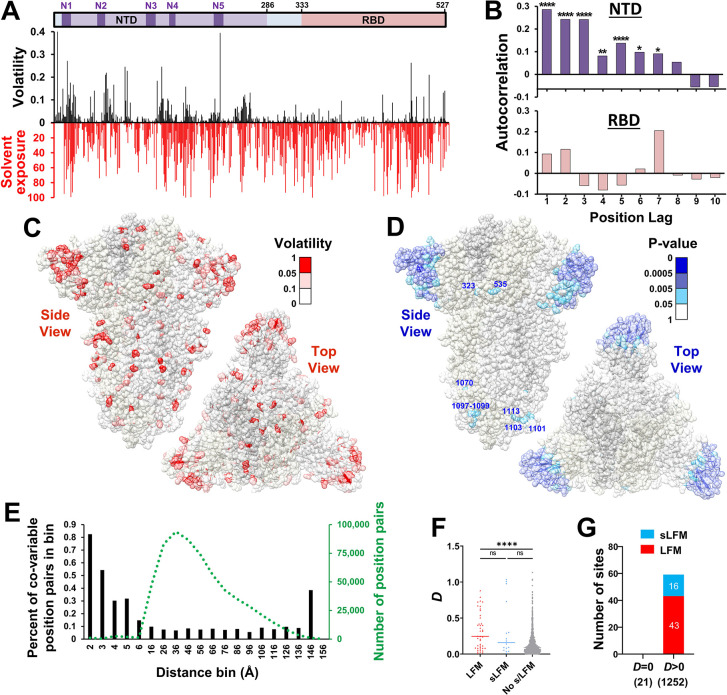
Fig 3. Spike positions in a volatile environment emerge as sites of sub/lineage-founder mutations.
(A) Volatility values for positions 1–527 were calculated for the SARS-CoV-2 BL group (black bars). Solvent exposure of the residues (based on spike structure 6ZGI) is expressed as a percent of the total solvent-accessible surface area of each residue (red bars). Positions unresolved on the structure (1–13 and 71–75) are assigned exposure values of 0. (B) Autocorrelation of volatility values for the NTD and RBD. Significance of the autocorrelation was calculated using the Spearman rank-order test: *, P<0.05; **, P<0.005; ***, P<0.0005; ****, P<0.00005. (C) Mapping of volatility values calculated for the BL group onto the spike trimer structure. (D) Results of a permutation test to identify positions with high volatility at their 10 closest neighbors on the trimer structure. Low P-values indicate sites with a high-volatility environment. Such sites located outside the NTD are labeled. (E) Fisher’s exact test was used to calculate the co-occurrence of a volatile state at any two positions of spike in the 114 clusters of the SARS-CoV-2 BL group. The distance between the closest atoms of any two residues was also calculated. Bars indicate the percent of position pairs with significant co-variability (P-value < 0.01) as a percent of all position pairs separated by the same distance range. The number of position pairs in each bin is indicated by the dotted green line. (F) The variable D describes for each position the total distance-weighted volatility at spike positions within 6Å on the trimer structure. D values are compared between positions with LFMs, sLFMs or no such mutations. (G) The number of positions that emerged with LFMs and sLFMs when the D value was zero or larger than zero. The number of positions in each subset is indicated in parentheses.