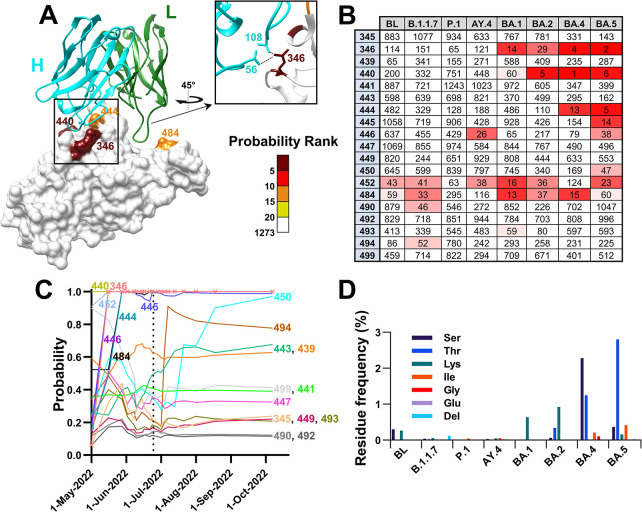
Fig 8. The mutation in subvariant BA.4.6 that conferred resistance to Cilgavimab is predicted well by volatility patterns in the parental BA.4 lineage.
(A) Mutation probabilities were calculated for all positions of spike using the baseline sequences of BA.4. Residues on the structure of the RBD in complex with Cilgavimab (PDB ID 7L7E) are colored by their probability ranks as indicated. (B) Probability ranks for contact sites of Cilgavimab, as calculated using the baseline sequences of the indicated lineages. (C) Sequences from the BA.4 baseline were indexed by their date of collection, divided into 50-sequence clusters, and mutation probability values were calculated for groups of increasing cluster numbers. Values are shown for all Cilgavimab contact sites. A vertical dashed line indicates the collection date of the first BA.4.6 sequence (D) Frequency of minority variants at position 346, expressed as a percent of all sequences in the baseline groups of the indicated lineages. Del, deletion event.