SUMMARY
Recreating tissue environments with precise control over mechanical, biochemical, and cellular organization is essential for next-generation tissue models for drug discovery, development studies, and the replication of disease environments. However, controlling these properties at cell-scale lengths remains challenging. Here, we report the development of printing approaches that leverage polyethylene glycol diacrylate (PEGDA) hydrogels containing photocaged oligonucleotides to spatially program material characteristics with non-destructive, non-ultraviolet light. We further integrate this system with a perfusion chamber to allow us to alter the composition of PEGDA hydrogels while retaining common light-activatable photocaged DNAs. We demonstrate that the hydrogels can capture DNA functionalized materials, including cells coated with complementary oligonucleotides with spatial control using biocompatible wavelengths. Overall, these materials open pathways to orthogonal capture of any DNA functionalized materials while not changing the sequences of the DNA.
Graphical Abstract

Recreating native tissues with precise control over mechanical properties and functionalities allows for more accurate tissue models. Mathis et al. report microscale formation of polyethylene glycol diacrylate hydrogels with mixed physicochemical properties, which incorporate photocaged DNA that can be activated across different mixed chemical and physical compositions.
INTRODUCTION
Native tissues are a complex and multifaceted environment that include multiple cell phenotypes and extracellular matrix (ECM) proteins arranged into hierarchical structures. The physical and chemical composition of the cellular microenvironment affects cell behaviors spanning migration and gene expression, while interactions between neighboring cells further modulate cell behaviors.1,2 Traditional methods of cell culture do not fully recreate tissue environments and do not accurately reflect the complexity of in vivo conditions altering cell responses.3,4 Current techniques for building tissue models often fall short. Microfluidic devices struggle with 3D complexity,5 bioprinting resolutions can be limited,6,7 and scaffolds have limited ability to incorporate multiple cell types. Even spheroids and organoids, while promising, are often not uniform in size and do not have fine structural control.8 To better understand cellular processes and imitate tissue structures for applications spanning drug delivery development, tissue engineering, and disease progression, we require methods for creating reproducible complex tissues with chemical, physical, and cellular heterogeneity reflective of physiology.
To achieve better in vitro models, hydrogels have proven to be a useful tool for replicating tissue structures due to their biocompatibility and ability to mimic the ECM that envelops cells in native tissues.9–11 Indeed, engineered hydrogels have allowed for directed cellular growth and differentiation.12–15 Nevertheless, a paramount challenge lies in imparting these hydrogel matrices with properties reflecting the cellular, mechanical, and chemical heterogeneity of tissues. To this end, hydrogels have been created with spatially controlled physical and biochemical properties, using photolithographic patterning techniques where gels are modified after synthesis.16–21 As an alternative to post-modification, hydrogels have been combined with solvent exchange systems to create unique structures with multifunctional behavior programmed into them using bottom-up synthetic approaches.22 In particular, polyethylene glycol diacrylate (PEGDA) hydrogels have played a prominent role in creating hydrogels with complex characteristics due to the ability to readily incorporate different functionalities (chemical and physical) into the structures.23–26 These materials have been used to create spatially heterogeneous chemical structures for 3D printing applications, highlighting their potential in tissue engineering applications.20,27–29
As a subset of hydrogels, materials with DNA appended to cross-links are particularly compelling as they allow one to utilize the DNA to program mechanical and chemical characteristics within hydrogels.30–35 For localized control over hydrogels, different DNA sequences have been placed in hydrogels using additive photolithographic methods16,36 and subtractive methods where the DNA is cleaved away through the use of photocleavable chemistries.30,37–41 Solvent exchange systems have been used to create PEGDA hydrogels with different DNA functionalities to create highly programmable materials with sequence-specific activities.42 In addition to imparting chemical and physical properties in hydrogels, DNA has been used to program specific cell interactions by coating cells with complementary DNA sequences to program cellular interactions and create 3D cell-dense assemblies.43–45 We recently demonstrated that photocaged oligonucleotides could be used to program specific cell interactions on cell monolayers with spatial control by activating DNA on cell surfaces with light.46 DNA-based cell localization has been extended to surfaces, including hydrogels,36 where cell placement was controlled using nucleic acids for capture of selected cell populations with spatial control. However, programming materials for orthogonal cell placement across chemically distinct regimens with high orthogonality and on-demand, maskless activation is difficult to achieve.
Here, we report the development of hydrogels with physiologically relevant stiffness that allow site-specific placement of cells and biomolecules. Specifically, we synthesize low-stiffness hydrogels that incorporate coumarin photocaged oligonucleotides and demonstrate that these materials can be repeatedly addressed with light to activate specific domains for recognition of complementary oligonucleotides. We demonstrate that these materials can be repeatedly activated for placement of cells. We then combine these materials with a solvent exchange system to print modular hydrogels with distinct chemical functionality that can be activated with light for complementary DNA recognition. Taken together, the materials reported allow for the creation of more complex in vitro models for applications spanning cell biology, tissue engineering, and drug discovery.
RESULTS AND DISCUSSION
Photocaged oligonucleotide-containing hydrogels for cell placement
To realize our goal of creating hydrogels with dynamic control over cell placement, we synthesized hydrogels that contained coumarin photocaged oligonucleotides. The coumarin groups block recognition of complementary DNA and can be selectively cleaved from the bases using ultraviolet (UV) and non-UV light (up to 500 nm) to activate the bases.47–50 Since we wanted to avoid unwanted uncaging of our DNA during hydrogel printing with UV light, we designed single-stranded DNA terminated with a thiol (DNA-SH; sequence 1; Table S1) that was partially complementary to DNA containing 5 photocaged bases (photocaged DNA [PC-DNA], sequence 2; Table S1), an amount of photocages needed to prevent unwanted hybridization (Figure 1A).46 The DNA-SH could then be cross-linked into a hydrogel and used to capture the photocaged DNA. The photocaged DNA then has a separate region of 20 bp that can be selectively activated with light to capture specific DNA sequences that could later be introduced to the hydrogels (Figure 1A). Electrophoretic gel shift assays showed that DNA-SH could hybridize to the photocaged DNA and that 425-nm light was able to uncage the DNA to allow for the additional capture of fluorophore-labeled DNA (sequence 3; Table S1; Figures 1B and S1). As a control, when we incubated the photocaged structures with non-complementary fluorophore-labeled DNA, no complementary binding was observed upon exposure to light; this shows that the capture of DNA was specific to DNA strands that were complementary to each other (Figure S2).
Figure 1. Capture and photoactivation of photocaged oligonucleotides.
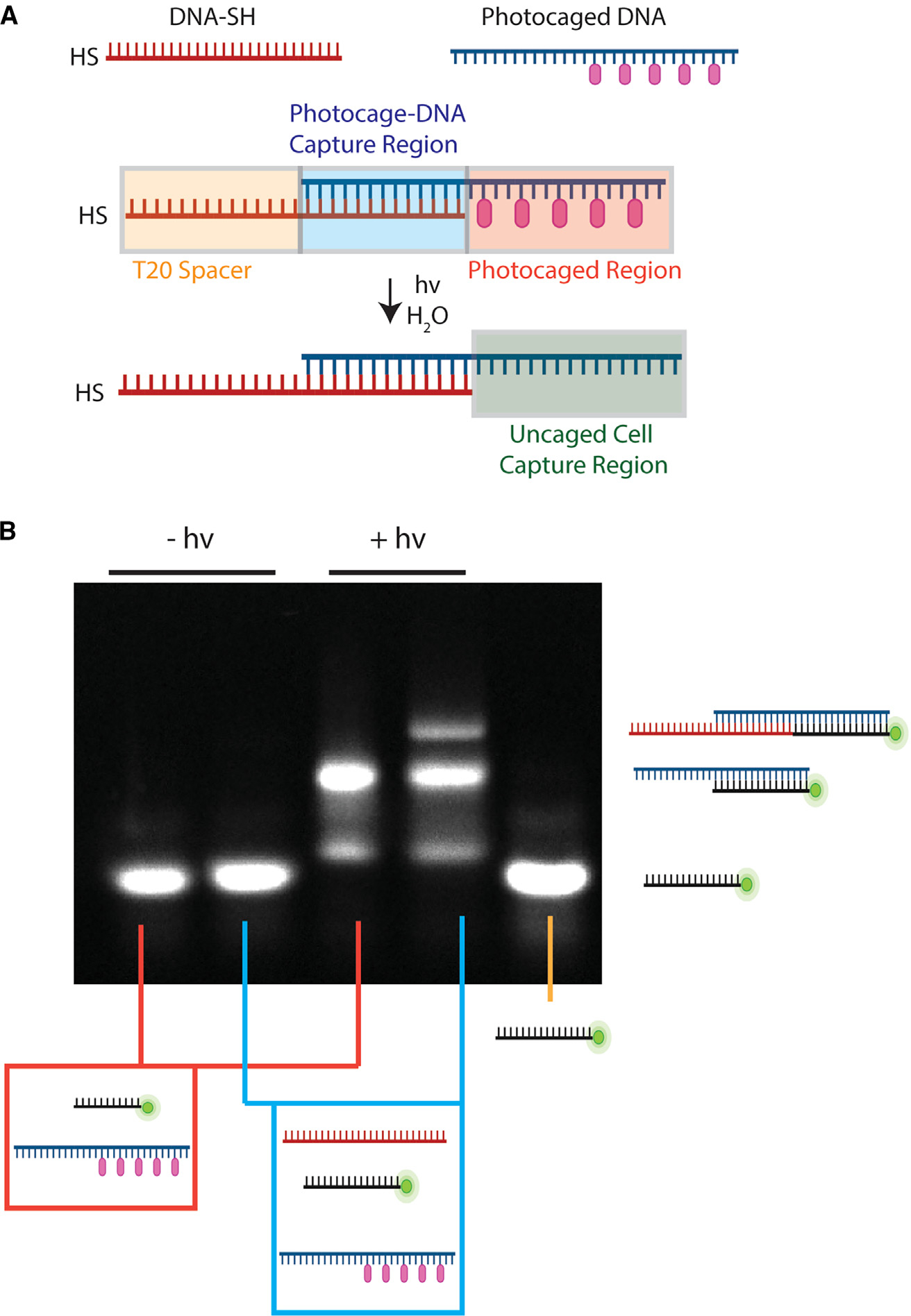
(A) Schematic of thiol DNA capturing the photocaged DNA that is activated upon exposure to light. Created with BioRender.
(B) Electrophoresis gel of the combinations of DNA. The first two lanes are DNA that is not exposed to light, and the second two lanes are DNA that is exposed to light. The first and third lanes are photocaged DNA and fluorescent complementary DNA. The second and fourth lanes also include DNA-SH. The fifth lane is fluorescent DNA only.
Our hydrogel synthesis scheme consisted of a two-step process. First, we synthesized PEGDA hydrogels using a thiolene chemistry that contained non-photocaged DNA, and second, we added photocaged DNA through partial hybridization with the SS-DNA cross-linked to our hydrogel (Figure 2A). For the synthesis of the hydrogels, methacrylated cover slips were coated with an aqueous solution of polyethylene glycol thiol (PEG-SH), PEGDA, and a water soluble photoinitiator, diphenyl (2,4,6-trimethylbenzoyl)-phosphine oxide nanoparticles (TPO-nanoparticles) along with DNA-SH mixed at stoichiometries guided by the work of others.51 We then polymerized this mixture by exposing it to 365-nm light. Importantly, we could tune the mechanical properties of the hydrogel by changing the amount of PEGDA in our system. Specifically, soft stiffnesses (~1–15 kPa) were achieved using concentrations ranging from (5–7 wt % PEGDA; Figure S3). Such soft materials are especially critical in many physiological environments, such as the lungs or meniscus, where mechanical properties contribute significantly to disease etiology.52,53
Figure 2. PEGDA hydrogels with photocaged oligonucleotides.
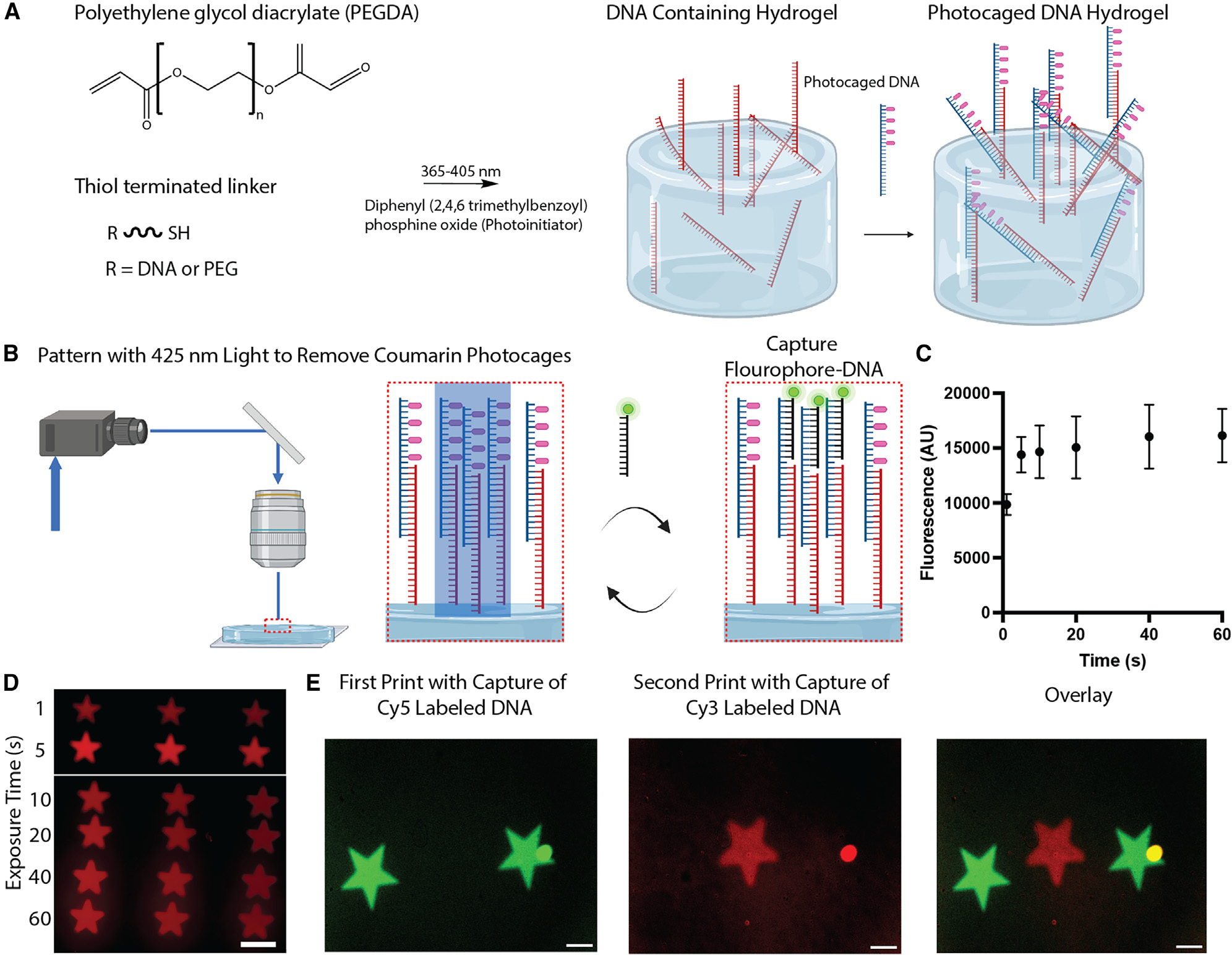
(A) Schematic detailing the steps in the initial hydrogel synthesis. Created with BioRender.
(B) Schematic of the photopatterning steps where photocaged DNA is deprotected and then incubated with complementary fluorophore-labeled SS-DNA. Created with BioRender.
(C) Graph showing the fluorescence as a function of the exposure time to 425-nm light.
(D) Fluorescence micrograph of patterned stars in hydrogels with different light exposure times.
(E) Fluorescence micrographs showing the capture of DNA strands labeled with different fluorophores.
All scale bars, 200 μm. Data presented as mean ± SEM.
After synthesizing the hydrogels with DNA, we incubated them with photocaged oligonucleotides. This step allowed us to add photoactivatable structures to our hydrogels that could be addressed with light repeatedly and in spatially localized areas while not being uncaged by light in the initial synthesis. To assess the removal of photocages from the DNA within the hydrogels, we exposed the hydrogels to light (425 nm) using a digital micromirror device (DMD) for different time periods (5, 10, 15, 30, and 60 s). The hydrogels were then incubated with fluorophore-labeled DNA (sequence 3; Table S1) complementary to the photocaged region (Figure 2B). Fluorescent micrographs show increasing fluorescence for areas exposed up to 20 s (Figures 2C and 2D). However, exposing the hydrogels beyond 20 s did not show a meaningful fluorescence increase, highlighting that activation of the DNA occurred quickly. Importantly, the hydrogels are redressable with repeat light exposures, allowing us to easily integrate additional functionalities. Specifically, we exposed one area of the hydrogel to light in the shape of a star, exposed to Cy5-labeled DNA, and then imaged the location of this pattern (Figure 2D). After rinsing the hydrogel, we could expose a nearby area on the hydrogel and remove photocages in this location. Incubation with Cy3-labeled DNA (sequence 4; Table S1) allowed us to visualize the second pattern area, which does not interfere with the first (Figure 2E). Only a small aggregate of material shows any significant overlap. Excluding the aggregate, the thresholded Mander’s coefficients are 0.009 and 0.109 for the two channels, indicating low colocalization.
Synthesis of localized hydrogels with mixed chemical properties and orthogonal light activation
Based on our hydrogel designs, we developed a solvent exchange system for printing localized hydrogels with kilopascal stiffnesses. To realize this, we integrated our hydrogel printing into a perfusion chamber and placed it on a microscope stage equipped with a DMD. The perfusion chamber allows us to dynamically change the composition of the polymerization materials, while the DMD allows us to perform spatially localized synthesis (Figure 3A). For our initial experiment, we focused on tuning the polymerization conditions to allow us to generate cell-scale features (~50 μm in width). We made solutions containing 7% (v/v) PEGDA, PEG-SH, and TPO. Upon perfusion and exposure to light, significant overpolymerization was observed regardless of the exposure time (Figure 3B). To reduce overpolymerization, we included Orange G, a water-soluble photoabsorber, used by several other groups in 3D printing of materials to enhance spatial control by absorbing UV light that is out of the focus region.54 Notably, our hydrogels are significantly softer than those traditionally printed in most applications, with PEGDA concentrations exceeding 30% (w/w). We created solutions consisting of 0.01%, 0.02%, and 0.05% Orange G and polymerized them with different exposure times. Our studies showed quick polymerization of the hydrogels with 0.05% Orange G with limited increases in feature thickness at longer exposure times (Figures S4 and S5). 0.01% Orange G showed rapid and poorly controlled polymerization at times as short as 10 s (Figures S4 and S5). We note that shorter exposure times did not show meaningful polymerization (and/or inconsistent polymerization), which is consistent with observations that sufficient energy is needed for initial gelation to occur before polymerization is observed.55 Based on our studies, we proceeded with 0.05% Orange G for polymerization studies.
Figure 3. Solvent exchange system for printing soft hydrogels on glass slides.

(A) Schematic of the flow chamber setup used in our printing procedure.
(B) Micrographs of hydrogels synthesized in the presence of different amounts of Orange G. Scale bars, 50 μm.
In addition to printing fidelity, we also assessed the mechanical properties of hydrogels to compare with bulk properties. Using atomic force microscopy (AFM), we probed the stiffness of hydrogels of 7.5% and 10% (w/w) PEGDA (Figure S6). Our measurements showed that the stiffness of our hydrogels was reduced in comparison with our bulk studies, which is consistent with literature observations that show that the mechanics of printed PEGDA hydrogels are not homogeneous across printed layers and that the layers farthest from the light source—the surface we probe with AFM—will be the softest.56 Despite the lower stiffnesses, our results still show that we can control the mechanical properties through PEGDA concentration.
Following our initial studies on polymerization conditions, we next proceeded to see whether we could synthesize hydrogels with low stiffness with mixed chemical properties. Importantly, we incorporated SS-DNA-SH for all polymerizations to allow for attachment of light-protected oligonucleotides in the post-processing of our hydrogels. In this study, we first flowed in a polymerization solution supplemented with rhodamine-PEG-thiol (Rh-PEG-SH) for easy visualization. We polymerized square patterns with different polymerization times of 20, 25, and 30 s. We then perfused in a PEGDA polymer solution supplemented with a biotin-PEG-thiol. We repeated our polymerization next to the original set of hydrogels. We then incubated the hydrogels with PC-DNA that again was partially complementary to the DNA-SH in the hydrogels to embed photocaged functionality. The gels were then exposed to light at their interface to activate only some of the DNA and then incubated with Cy5-labeled DNA that was complementary to the decaged portion of the PC-DNA. Micrographs show that two distinct hydrogels are placed next to each other for all patterns (Figure 4), while fluorescence micrographs show that only one of the gels contains the Rh-PEG-SH functionality (Figures 4B and 4C). In addition, localized Cy5 fluorescence was observed across the interface of the two gels, where they were exposed to 425-nm light.
Figure 4. Multifunctional hydrogel with orthogonal light activation of DNA.

(A) Schematic of the printing process for including different functional domains in the hydrogels along with uncaging of DNA in hydrogels.
(B) Micrographs showing different hydrogel exposure times along with different DNA deprotection times.
(C) Magnified fluorescence micrograph images of one of the hydrogels. Scale bar, 400 μm.
Patterning of cells on hydrogels using local photoactivation of DNA
Next, we assessed whether our photocaged DNA in our hydrogels could be used to localize cells. For these experiments, we first synthesized thin PEGDA/PEG-SH hydrogels with DNA-SH and then incubated the hydrogels with PC-DNA for light activation. The gels were then patterned for 60 s with 425 nm and then incubated with fluorophore-labeled cells coated with DNA complementary to the hydrogel (sequence 5; Table S1). Cells were added to excess to ensure that there were fast interactions with the surface (incubation time of ~5 min) and that all pattern locations had cells. After removing excess cells and rinsing with PBS, repeated patterns of cells configured into star shapes or annuli were observed (Figure 5). Cells on the star patterns were imaged 4 days later to confirm that cells were still present and to assess pattern retention. 4 days later, significant pattern degradation was observed, which is expected, as DNA will be degraded (Figure S7). As controls, cells without complementary DNA did not stick to defined patterns (Figure S8A). Subsequent incubation with Cy3-labeled DNA confirmed that localized uncaging was achieved (Figure S8B). Overall, our experiments show that photocaged DNA hydrogels are a viable method for creating on-demand cell placement into arbitrary shapes.
Figure 5. Fluorescence micrographs of cells patterned using photocaged DNA.

(A and B) Fluorophore-labeled cells patterned into the shape of stars (scale bar, 500 μm).
(C) Fluorophore-labeled cells in the shape of annuli (scale bar, 200 μm).
In summary, we presented a pathway to orthogonal control over material properties and cell population localization using photocaged oligonucleotides. By combining these hydrogels with proteins, peptides, and different PEGDA compositions, we can readily create hydrogels with highly programmable properties for mimicking cellular environments. Importantly, combining this technology with multiplexed automated printing would allow one to readily introduce cellular, protein, and mechanical attributes to the material on demand. Importantly, the use of coumarin protecting groups allows for all this to be done in cell-compatible fashions due to the lack of UV exposure and intrinsic cell compatibility of non-targeted small nucleic acids. Collectively, these materials have the potential to advance developmental biology, drug discovery, and 3D printing.
EXPERIMENTAL PROCEDURES
Resource availability
Lead contact
Requests for further information, resources, or materials should be directed to and will be fulfilled by the lead contact, Brian Meckes (brian.meckes@unt.edu), upon reasonable request.
Materials availability
No new unique materials were generated by this study.
Data and code availability
Data included in this manuscript and the supplemental information will be made available by the lead contact upon reasonable request.
Safety
No unexpected, new, or significant hazards are associated with this work. Human cell lines were obtained from authorized sources and used according to institutional biosafety protocols, and all chemical handling followed established laboratory safety practices with appropriate personal protective equipment worn.
Coverslip preparation
22 × 22 mm square glass coverslips were submerged in ethanol (Fisher Chemical) and sonicated for 30 min. The coverslips were dried with compressed air and placed into two 6-well plates. Next, the glass coverslips were plasma etched with oxygen in a PE-25 series plasma system (Plasma Etch) for 15 min to activate the surface. The coverslips were placed in a 20-mL solution consisting of 1% acetic acid (v/v), 2% 3-(methacryloyloxy)propyl]trimethoxysilane (TMSPMA) (v/v), and 97% ethanol (v/v) for 5 min with the plasma-etched side facing upward, as reported previously.57 The coverslips were then rinsed with distilled water and dried with compressed air. Then, the coverslips were placed on microscope slides and placed in the Thermocenter (Salvis) oven to dry at 120°C for 1 h. The coverslips were stored in the desiccator for use within 1–2 days.
Cell culture and staining
NM2C5 cells (ATCC, CRL-2918) were maintained in DMEM with 4.5 g/L glucose, L-glutamine, and sodium pyruvate (Corning), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS). Cells were cultured at 37°C and 5% CO2. Cells were passaged at 80% confluency, and the medium was changed every 2–3 days. Live-cell staining was done with CellTracker (Invitrogen). CellTracker dye was resolubilized with dimethyl sulfoxide (DMSO) to a 10 mM stock concentration and diluted at 1:1,000 with complete medium. This working solution was added to the cells for 45 min at 37°C. Cells were imaged using an Olympus BX63 microscope with the relevant filter sets. Cell imaging was completed on the same day for each experiment, using the same microscope settings.
DNA synthesis
All DNA monomers and reagents were purchased from Glen Research. DNA was synthesized with the Expedite synthesizer (Biolytic) following the manufacturer’s standard protocols. We used standard bases along with the photoreactive coumarin-protected guanine (5′-dimethoxytrityl-N2-(4-isopropylphenoxyacetyl)-O6-[[7-(diethylamino)coumarin-4-yl]-methyl]-2′-deoxyguanosine, 3′-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite). All oligonucleotides were synthesized with the dimethoxytrityl-on for purification. Oligonucleotides were deprotected using water saturated with ammonium hydroxide (28%–32%, Fisher Chemicals) for 24 h at room temperature and then dried under air (Organomation). Oligonucleotides were then purified using a Glen-Pac DNA purification column (Glen Research), following the manufacturers protocols. Samples were subsequently lyophilized (Labconco, −105°C) and reconstituted to target dilutions in water. For TCO-modified oligonucleotides, oligos were synthesized with a 5′ C6 amino group (6-(4,4′-dimethoxy-4″-methylsulfonyl-tritylamino)hexyl-(2-cyanoethyl)-(N,N-diisopropyl)-phosphoramidite). After cleavage of the trityl group and purification, the oligonucleotides were incubated with transcyclooctene-PEG6-N-hydroxysuccinimide ester (TCO-PEG6-NHS ester; Click Chemistry Tools; prepared in anhydrous DMSO) with 10:1 (mol:mol) of DNA synthesized in 10 mM HEPES-buffered saline (pH 8.4); it is important to add DNA directly to the TCO-PEG6-NHS ester at the required concentrations to prevent unwanted hydrolysis. Oligonucleotides were then purified on a GLEN Gel-Pak desalting column (Glen Research), following the manufacturer protocol.
Hydrogel synthesis for light patterning experiments
A 7.2% PEGDA hydrogel was synthesized by creating aqueous mixtures of PEGDA (Sigma-Aldrich, 7.2% [v/v]), 60 μL water-soluble TPO-nanoparticles (Sigma-Aldrich, 10 mg/mL), 1 K PEG-thiol (Creative Pegworks, 10 mg/mL in H2O), and DNA-SH (100 μM). All stock solutions prepared in DI H2O. Next, 45 μL of the solution was added to the plasma-etched glass coverslips, and a new coverslip was placed on top of the solution, creating an even layer of hydrogel. The coverslips with the solution were then exposed to 365-nm light for 10 min to form a hydrogel. Following this, the coverslip on top of the hydrogel was removed. Photoprotected Adaptor DNA (PC-DNA, sequence 2; Table S1; 1 μM) was placed in PBS and shaken for 5 min. The hydrogel was protected from light during these steps to prevent unwanted degradation. They were then rinsed with 1 × PBS 3 times, for 5 min during each rinse. 425-nm LEDs (ThorLabs, power of 1,445 mW) for different time intervals using a DMD (Mightex) attached to a microscope (BX63, Olympus). The cells were incubated with 1 μM Cy5-labeled complementary DNA for 2 min and then rinsed with 1 × phosphate-buffered saline (PBS; Hyclone Cytiva). The PBS contained 144 mg/L potassium phosphate monobasic, 9,000 mg/L sodium chloride, and 795 mg/L sodium phosphate dibasic.
Multifunctional hydrogel synthesis
For multifunctional hydrogels, two 9.35% PEGDA solutions were synthesized by creating aqueous solutions of PEGDA, TPO-nanoparticles (10 mg/mL in H2O stock), PEG-SH (10 mg/mL in H2O), Orange G (5 μg/mL, 0.05 wt %), and 4-methoxyphenol (MET-PH) (25 μM in H2O). Following this, either Rh-PEG-SH (200 nM) or biotin thiol (200 nm) was added to the solution. Next, to make the hydrogel, a perfusion chamber (Werner Instruments) was used. Before use, the perfusion chamber was prepared by applying a small amount of vacuum grease to the square outline at the bottom of the chamber. Once the grease was applied, a normal coverslip was placed on the grease to ensure that the solution would not leak when placed through the chamber. Then, methacrylated coverslips were placed on top, with the methacrylated surface pointing down. The magnet was then placed on top of the perfusion chamber to complete the seal. Solution was injected into the perfusion chamber. Areas of the perfusion chamber were then selectively exposed to varying times of 365-nm light using the Mightex projector on an Olympus BX63 microscope. Next, the perfusion chamber was rinsed with deionized (DI) H2O. The biotin thiol hydrogel solution was then injected into the perfusion chamber. A square pattern was then exposed to 365-nm light at the same time points. Following this, the perfusion chamber was once again rinsed with DI H2O and patted dry with chemical wipes. Last, the perfusion chamber was disassembled, and the coverslip with the polymerized hydrogels was imaged.
For experiments with photocaged DNA, 100 μM DNA-SH was added to each of the prior hydrogel solutions. After the hydrogels were created, the gels were incubated with 5 μM of PC-DNA. Sections of the gels were then patterned with 425-nm light using the DMD. After patterning, the gel was rinsed with 1 × PBS 3 times in the 6 well plate. Then, fluorescent complementary DNA (1 μM in PBS) was added to the well for 1 min and then rinsed with 1 × PBS 3 times.
Perfusion chamber experiments with Orange G
Pre-polymer ink batches were prepared as aqueous solutions of 6% PEGDA (v/v), TPO-nanoparticles (10 mg/mL), 1 K PEG-SH (10 mg/mL), Rh-PEG-SH (200 nM), and Orange G (0.01%, 0.02%, and 0.05% [w/w]). The ink was flowed into a perfusion chamber containing an TMSPMA-functionalized glass coverslip, and microscopic patterns were projected onto the surface by a 365-nm light for 10, 15, 20, and 30 s (as described above). Following photopatterning, the coverslips were rinsed five times with DI water by dilution and imaged.
Mechanical testing on bulk hydrogels
Pre-polymer inks were prepared as aqueous solutions at a working concentration of TPO-nanoparticles (4 mg/mL), PEG-SH (10 mg/mL), and PEGDA at concentrations of 5%, 5.5%, 6%, 6.5%, and 7% (v/v). The ink solutions were pipetted into 12-well cell culture plates, and a TMSPMA-functionalized microscope slide was placed on top of each filled well. The 12-well plate was then placed into a UV incubator at a wavelength of 365 nm for 20 min. After carefully removing the bulk hydrogels from the plates, they were stored in water overnight.
The average elastic modulus was determined through mechanical testing.58 Metal beads with diameters of 4.5 mm, 5.5 mm, 6 mm, 7 mm, and 8 mm were placed on top of each gel. A picture leveled at the surface of the gel was taken, and the indentation depth of the bead was measured with ImageJ. Using Young’s modulus formula, the elastic modulus of each gel was calculated. At each tested concentration of PEGDA, the elastic modulus of three gels was measured and averaged to determine the average elastic modulus.
AFM mechanical testing
Hydrogels with defined chemical compositions were synthesized as described above. The glass coverslips were mounted onto metal AFM pucks (Ted Pella), and the mechanics were assessed using an atomic force microscope (Multimode III, Digital Instruments). All measurements were performed in hydrated environments using a fluid cell (Digital Instruments). AFM cantilevers with 3.5 μm silicon beads mounted to the tip and nominal spring constants of 0.08 N/m (Nanoandmore) were used in all experiments. Deflection sensitivity was assessed by probing a glass substrate that was treated as non-deformable for these experiments. Force ramps were acquired at indentation rates of 2 μm/s. Force curves were evaluated using Nanoscope Analysis (Bruker) using 10%–70% of the force curve and Hertzian spherical indentation model.
Cell patterning experiments
DNA is attached to the cell membrane through reactions involving methyltetrazine (MTET) sulfo-NHS ester, as we described previously.46 Briefly, the NHS ester group reacts with primary amines of proteins on the cell surface, and the MTET reacts with a TCO attached to the DNA, binding the DNA to the cell surface. A maximum of 600,000 cells were suspended in 100 μL of 1 × PBS. A 200 mM stock solution of MTET-sulfo-NHS ester (Click Chemistry Tools) in anhydrous DMSO was added to the cells at 1 mM in 10 mM HEPES (pH 8.4) with 150 mM NaCl. After 5 min on a shaker at 30°C, the cells were rinsed twice by centrifuging at 250 × g for 5 min, removing the supernatant, and adding 100 μL of PBS. TCO-DNA was added to the cells at 100 μM in medium, left for 5 min at room temperature, and rinsed twice by centrifuging. The cells were then resuspended with 100 μL of 1 × PBS. The cells were added straight onto patterned hydrogels and left without shaking for 5 min. Then, the excess cells were rinsed away with 1 × PBS.
Supplementary Material
Highlights.
Hydrogels are spatially activated with light to capture specific DNA sequences
Hydrogels have different chemical functionalities in defined regions
Photocaged DNA in hydrogels allows for spatial activation of functionality
Cell placement controlled through light activation of photocaged DNA in hydrogels
ACKNOWLEDGMENTS
The research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under awards R21GM141563 and R35GM150577. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.xcrp.2024.101922.
REFERENCES
- 1.Chung S, Sudo R, Mack PJ, Wan C-R, Vickerman V, and Kamm RD (2009). Cell migration into scaffolds under co-culture conditions in a microfluidic platform. Lab Chip 9, 269–275. [DOI] [PubMed] [Google Scholar]
- 2.Yang H, Borg TK, Ma Z, Xu M, Wetzel G, Saraf LV, Markwald R, Runyan RB, and Gao BZ (2016). Biochip-based study of unidirectional mitochondrial transfer from stem cells to myocytes via tunneling nanotubes. Biofabrication 8, 015012. [DOI] [PubMed] [Google Scholar]
- 3.Fontoura JC, Viezzer C, Dos Santos FG, Ligabue RA, Weinlich R, Puga RD, Antonow D, Severino P, and Bonorino C (2020). Comparison of 2D and 3D cell culture models for cell growth, gene expression and drug resistance. Mater. Sci. Eng., C 107, 110264. [DOI] [PubMed] [Google Scholar]
- 4.Pampaloni F, Reynaud EG, and Stelzer EHK (2007). The third dimension bridges the gap between cell culture and live tissue. Nat. Rev. Mol. Cell Biol. 8, 839–845. [DOI] [PubMed] [Google Scholar]
- 5.Coluccio ML, Perozziello G, Malara N, Parrotta E, Zhang P, Gentile F, Limongi T, Raj PM, Cuda G, Candeloro P, and Di Fabrizio E (2019). Microfluidic platforms for cell cultures and investigations. Microelectron. Eng. 208, 14–28. [Google Scholar]
- 6.Nikolova MP, and Chavali MS (2019). Recent advances in biomaterials for 3D scaffolds: A review. Bioact. Mater. 4, 271–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JM, Ng WL, and Yeong WY (2019). Resolution and shape in bioprinting: Strategizing towards complex tissue and organ printing. Appl. Phys. Rev. 6. [Google Scholar]
- 8.Lancaster MA, and Knoblich JA (2014). Organogenesis in a dish: modeling development and disease using organoid technologies. Science 345, 1247125. [DOI] [PubMed] [Google Scholar]
- 9.Naahidi S, Jafari M, Logan M, Wang Y, Yuan Y, Bae H, Dixon B, and Chen P (2017). Biocompatibility of hydrogel-based scaffolds for tissue engineering applications. Biotechnol. Adv. 35, 530–544. [DOI] [PubMed] [Google Scholar]
- 10.Wechsler ME, Rao VV, Borelli AN, and Anseth KS (2021). Engineering the MSC secretome: a hydrogel focused approach. Adv. Healthcare Mater. 10, 2001948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geckil H, Xu F, Zhang X, Moon S, and Demirci U (2010). Engineering hydrogels as extracellular matrix mimics. Nanomedicine 5, 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bryant SJ, Arthur JA, and Anseth KS (2005). Incorporation of tissue-specific molecules alters chondrocyte metabolism and gene expression in photocrosslinked hydrogels. Acta Biomater. 1, 243–252. [DOI] [PubMed] [Google Scholar]
- 13.Mabry KM, Payne SZ, and Anseth KS (2016). Microarray analyses to quantify advantages of 2D and 3D hydrogel culture systems in maintaining the native valvular interstitial cell phenotype. Biomaterials 74, 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hynd MR, Frampton JP, Dowell-Mesfin N, Turner JN, and Shain W (2007). Directed cell growth on protein-functionalized hydrogel surfaces. J. Neurosci. Methods 162, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin S, and Cao Y (2021). Hydrogels for large-scale expansion of stem cells. Acta Biomater. 128, 1–20. [DOI] [PubMed] [Google Scholar]
- 16.Ramani N, Figg CA, Anderson AJ, Winegar PH, Oh E, Ebrahimi SB, Samanta D, and Mirkin CA (2023). Spatially-Encoding Hydrogels with DNA to Control Cell Signaling. Adv. Mater. 35, 2301086. [DOI] [PubMed] [Google Scholar]
- 17.Nelson BR, Kirkpatrick BE, Miksch CE, Davidson MD, Skillin NP, Hach GK, Khang A, Hummel SN, Fairbanks BD, Burdick JA, et al. (2023). Photoinduced dithiolane crosslinking for multiresponsive dynamic hydrogels. Adv. Mater. 2211209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stowers RS, Allen SC, and Suggs LJ (2015). Dynamic Phototuning of 3D Hydrogel Stiffness, 112 (Proceedings of the National Academy of Sciences; ), pp. 1953–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong DY, Griffin DR, Reed J, and Kasko AM (2010). Photodegradable hydrogels to generate positive and negative features over multiple length scales. Macromolecules 43, 2824–2831. [Google Scholar]
- 20.Nemir S, Hayenga HN, and West JL (2010). PEGDA hydrogels with patterned elasticity: Novel tools for the study of cell response to substrate rigidity. Biotechnol. Bioeng. 105, 636–644. [DOI] [PubMed] [Google Scholar]
- 21.Oh E, Meckes B, Chang J, Shin D, and Mirkin CA (2022). Controlled Glioma Cell Migration and Confinement Using Biomimetic-Patterned Hydrogels. Advanced NanoBiomed Research; 2, 2100131. [Google Scholar]
- 22.Xie X, Xu Z, Yu X, Jiang H, Li H, and Feng W (2023). Liquid-in-liquid printing of 3D and mechanically tunable conductive hydrogels. Nat. Commun. 14, 4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunckler MD, Medina JD, Coronel MM, Weaver JD, Stabler CL, and García AJ (2019). Linkage Groups within Thiol–Ene Photoclickable PEG Hydrogels Control In Vivo Stability. Adv. Healthcare Mater. 8, 1900371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuchiara MP, Allen ACB, Chen TM, Miller JS, and West JL (2010). Multilayer microfluidic PEGDA hydrogels. Biomaterials 31, 5491–5497. [DOI] [PubMed] [Google Scholar]
- 25.Zhu J, Tang C, Kottke-Marchant K, and Marchant RE (2009). Design and synthesis of biomimetic hydrogel scaffolds with controlled organization of cyclic RGD peptides. Bioconjugate Chem. 20, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gonzalez AL, Gobin AS, West JL, McIntire LV, and Smith CW (2004). Integrin interactions with immobilized peptides in polyethylene glycol diacrylate hydrogels. Tissue Eng. 10, 1775–1786. [DOI] [PubMed] [Google Scholar]
- 27.Hao Y, and Lin CC (2014). Degradable thiol-acrylate hydrogels as tunable matrices for three-dimensional hepatic culture. J. Biomed. Mater. Res. 102, 3813–3827. [DOI] [PubMed] [Google Scholar]
- 28.Teng D. y., Wu Z. m., Zhang X. g., Wang Y. x., Zheng C, Wang Z, and Li C. x. (2010). Synthesis and characterization of in situ cross-linked hydrogel based on self-assembly of thiol-modified chitosan with PEG diacrylate using Michael type addition. Polymer 51, 639–646. [Google Scholar]
- 29.Vanderhooft JL, Mann BK, and Prestwich GD (2007). Synthesis and characterization of novel thiol-reactive poly (ethylene glycol) cross-linkers for extracellular-matrix-mimetic biomaterials. Biomacromolecules 8, 2883–2889. [DOI] [PubMed] [Google Scholar]
- 30.Dorsey PJ, Rubanov M, Wang W, and Schulman R (2019). Digital maskless photolithographic patterning of DNA-functionalized poly (ethylene glycol) diacrylate hydrogels with visible light enabling photodirected release of oligonucleotides. ACS Macro Lett. 8, 1133–1140. [DOI] [PubMed] [Google Scholar]
- 31.Shi R, Fern J, Xu W, Jia S, Huang Q, Pahapale G, Schulman R, and Gracias DH (2020). Multicomponent DNA polymerization motor gels. Small 16, 2002946. [DOI] [PubMed] [Google Scholar]
- 32.Morya V, Walia S, Mandal BB, Ghoroi C, and Bhatia D (2020). Functional DNA based hydrogels: Development, properties and biological applications. ACS Biomater. Sci. Eng. 6, 6021–6035. [DOI] [PubMed] [Google Scholar]
- 33.Athanasiadou D, Meshry N, Monteiro NG, Ervolino-Silva AC, Chan RL, McCulloch CA, Okamoto R, and Carneiro KM (2023). DNA hydrogels for bone regeneration. In Proceedings of the National Academy of Sciences, 120Proceedings of the National Academy of Sciences, pp. e2220565120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C, Chen P, Shao Y, Zhou X, Wu Y, Yang Z, Li Z, Weil T, and Liu D (2015). A Writable Polypeptide–DNA Hydrogel with Rationally Designed Multi-modification Sites. Small 11, 1138–1143. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Chao J, Liu H, Su S, Wang L, Huang W, Willner I, and Fan C (2017). Clamped hybridization chain reactions for the self-assembly of patterned DNA hydrogels. Angew. Chem., Int. Ed. Engl. 56, 2171–2175. [DOI] [PubMed] [Google Scholar]
- 36.Prahl LS, Porter CM, Liu J, Viola JM, and Hughes AJ (2023). Independent control over cell patterning and adhesion on hydrogel substrates for tissue interface mechanobiology. iScience 26, 106657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kahn JS, Hu Y, and Willner I (2017). Stimuli-responsive DNA-based hydrogels: from basic principles to applications. Acc. Chem. Res. 50, 680–690. [DOI] [PubMed] [Google Scholar]
- 38.Wei Y, Wang K, Luo S, Li F, Zuo X, Fan C, and Li Q (2022). Programmable DNA hydrogels as Artificial extracellular matrix. Small 18, 2107640. [DOI] [PubMed] [Google Scholar]
- 39.Huang F, Chen M, Zhou Z, Duan R, Xia F, and Willner I (2021). Spatiotemporal patterning of photoresponsive DNA-based hydrogels to tune local cell responses. Nat. Commun. 12, 2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cangialosi A, Yoon C, Liu J, Huang Q, Guo J, Nguyen TD, Gracias DH, and Schulman R (2017). DNA sequence–directed shape change of photopatterned hydrogels via high-degree swelling. Science 357, 1126–1130. [DOI] [PubMed] [Google Scholar]
- 41.O’Hagan MP, Duan Z, Huang F, Laps S, Dong J, Xia F, and Willner I (2023). Photocleavable Ortho-Nitrobenzyl-Protected DNA Architectures and Their Applications. Chem. Rev. 123, 6839–6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubanov M, Cole J, Lee H-J, Soto Cordova LG, Chen Z, Gonzalez E, and Schulman R (2024). Multi-domain automated patterning of DNA-functionalized hydrogels. PLoS One 19, e0295923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandra RA, Douglas ES, Mathies RA, Bertozzi CR, and Francis MB (2006). Programmable cell adhesion encoded by DNA hybridization. Angew. Chem., Int. Ed. Engl. 45, 896–901. [DOI] [PubMed] [Google Scholar]
- 44.Gartner ZJ, and Bertozzi CR (2009). Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. In Proceedings of the National Academy of Sciences, 106Proceedings of the National Academy of Sciences, pp. 4606–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song P, Shen J, Ye D, Dong B, Wang F, Pei H, Wang J, Shi J, Wang L, Xue W, et al. (2020). Programming bulk enzyme heterojunctions for biosensor development with tetrahedral DNA framework. Nat. Commun. 11, 838–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathis K, Kohon AI, Black S, and Meckes B (2023). Light-Controlled Cell–Cell Assembly Using Photocaged Oligonucleotides. ACS Mater. Au 3, 386–393. 10.1021/acsmaterialsau.3c00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang X, Richards JL, Peritz AE, and Dmochowski IJ (2005). Photoregulation of DNA polymerase I (Klenow) with caged fluorescent oligodeoxynucleotides. Bioorg. Med. Chem. Lett. 15, 5303–5306. [DOI] [PubMed] [Google Scholar]
- 48.Lusic H, Young DD, Lively MO, and Deiters A (2007). Photochemical DNA activation. Org. Lett. 9, 1903–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodrigues-Correia A, Weyel XMM, and Heckel A (2013). Four levels of wavelength-selective uncaging for oligonucleotides. Org. Lett. 15, 5500–5503. [DOI] [PubMed] [Google Scholar]
- 50.Menge C, and Heckel A (2011). Coumarin-caged dG for improved wavelength-selective uncaging of DNA. Org. Lett. 13, 4620–4623. [DOI] [PubMed] [Google Scholar]
- 51.Zhang X, Ding S, Magoline J, Ivankin A, and Mirkin CA (2022). Photopolymerized Features via Beam Pen Lithography as a Novel Tool for the Generation of Large Area Protein Micropatterns. Small 18, 2105998. [DOI] [PubMed] [Google Scholar]
- 52.Kwok J, Grogan S, Meckes B, Arce F, Lal R, and D’Lima D (2014). Atomic force microscopy reveals age-dependent changes in nanomechanical properties of the extracellular matrix of native human menisci: implications for joint degeneration and osteoarthritis. Nanomedicine 10, 1777–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lai-Fook SJ, and Hyatt RE (2000). Effects of age on elastic moduli of human lungs. J. Appl. Physiol. 89, 163–168. [DOI] [PubMed] [Google Scholar]
- 54.Mau R, Nazir J, John S, and Seitz H (2019). Preliminary study on 3D printing of PEGDA hydrogels for frontal sinus implants using digital light processing (DLP). Current Directions in Biomedical Engineering 5, 249–252. [Google Scholar]
- 55.Jacobs PF (1992). Fundamentals of Stereolithography. [Google Scholar]
- 56.Hakim Khalili M, Panchal V, Dulebo A, Hawi S, Zhang R, Wilson S, Dossi E, Goel S, Impey SA, and Aria AI (2023). Mechanical Behavior of 3D Printed Poly (ethylene glycol) Diacrylate Hydrogels in Hydrated Conditions Investigated Using Atomic Force Microscopy. ACS Appl. Polym. Mater. 5, 3034–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mandal K, Wang I, Vitiello E, Orellana LAC, and Balland M (2014). Cell dipole behaviour revealed by ECM sub-cellular geometry. Nat. Commun. 5, 5749. [DOI] [PubMed] [Google Scholar]
- 58.Gandin A, Murugesan Y, Torresan V, Ulliana L, Citron A, Contessotto P, Battilana G, Panciera T, Ventre M, Netti AP, et al. (2021). Simple yet effective methods to probe hydrogel stiffness for mechanobiology. Sci. Rep. 11, 22668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in this manuscript and the supplemental information will be made available by the lead contact upon reasonable request.


