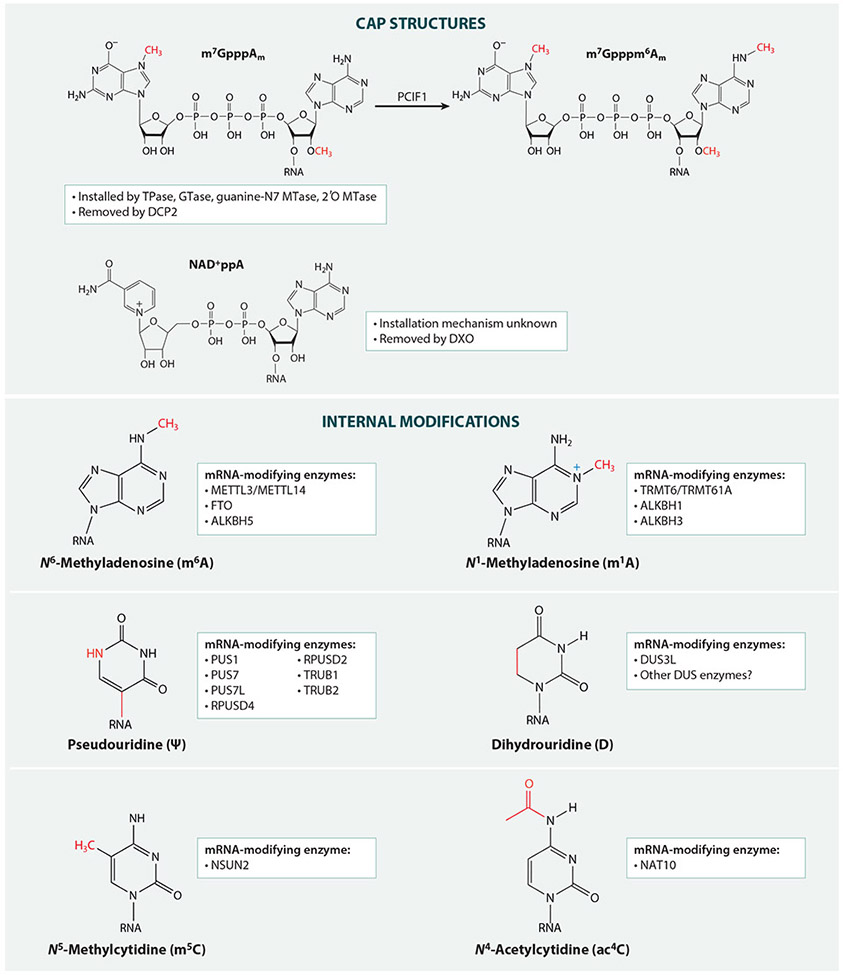
Figure 1.
Chemical structures of the modifications described in this review, as well as known enzymes that install and remove these modifications specifically on mRNA. Abbreviations: Ψ, pseudouridine; ac4C, N4-acetylcytidine; ALKBH, alkB homolog; D, dihydrouridine; DCP2, decapping mRNA 2; DUS, dihydrouridine synthase; DXO, decapping exoribonuclease; FTO, alpha-ketoglutarate dependent dioxygenase; GTase, guanylyl transferase; m1A, N1-methyladenosine; m5C, N5-methylcytidine; m6A, N6-methyladenosine; METTL, methyltransferase like; MTase, methyltransferase; NAD+ppA, NAD+ cap structure with 2′-O-methylated adenosine; NAT10, nuclear acetyltransferase 10; NSUN2, NOP2/Sun RNA methyltransferase 2; PCIF1, phosphorylated CTD interacting factor 1; PUS, pseudouridine synthase; RPUS, RNA pseudouridine synthase; TPase, RNA triphosphatase; TRMT, tRNA methyltransferase; TRUB, TruB pseudouridine synthase family.