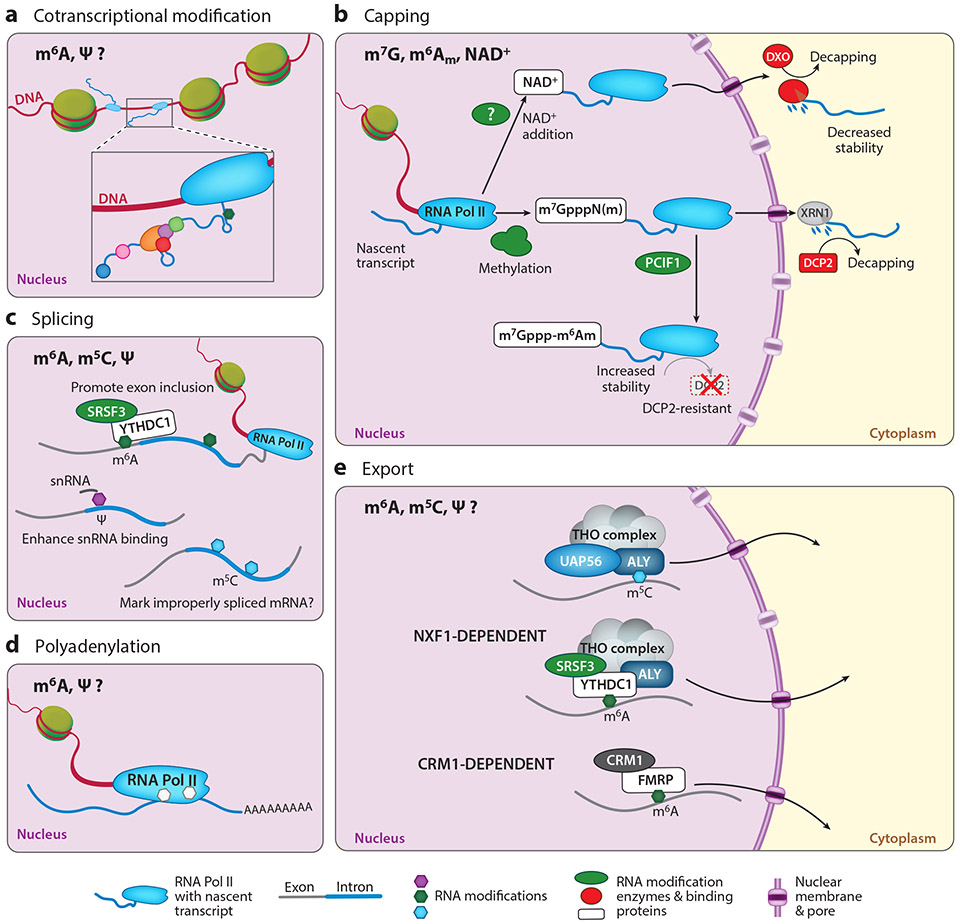
Figure 3.
mRNA modifications can be installed cotranscriptionally and regulate multiple steps in nuclear processing and export. (a) Some modifications are installed cotranscriptionally via direct recruitment of modification enzymes to RNA Pol II as it synthesizes nascent transcripts. (b) mRNA capping involves multiple modifications of the 5′ end of transcripts, which can directly modulate mRNA stability by altering susceptibility to decapping and degradation enzymes. (c) Splicing also often occurs cotranscriptionally and has been shown to be regulated by multiple modifications that can alter mRNA–snRNA interactions, recruit proteins that regulate exon inclusion, or mark improperly spliced transcripts. (d) Less is known about how mRNA modifications influence polyadenylation, but both m6A and Ψ may play important roles. (e) RNA modifications likely also influence the nuclear export of properly processed transcripts through both the NXF1- and CRM1-dependent pathways. Abbreviations: Ψ, pseudouridine; ALY, Aly/REF export factor; CRM1, exportin 1; DCP2, decapping mRNA 2; DXO, decapping exoribonuclease; FMRP, fragile X messenger ribonucleoprotein; m5C, N5-methylcytidine; m6A, N6-methyladenosine; m6Am, N6-2′-O-dimethyladenosine; m7G, N7-methylguanosine; NAD+, nicotinamide-adenine dinucleotide; NXF1, nuclear RNA export factor 1; PCIF1, phosphorylated CTD interacting factor 1; RNA Pol II, RNA polymerase II; snRNA, small nuclear RNA; SRSF3, serine and arginine rich splicing factor 3; THO, THO nuclear export complex; UAP56, DExD-box helicase 39B; XRN1, 5′–3′ exoribonuclease 1; YTHDC1, YTH domain containing protein 1.