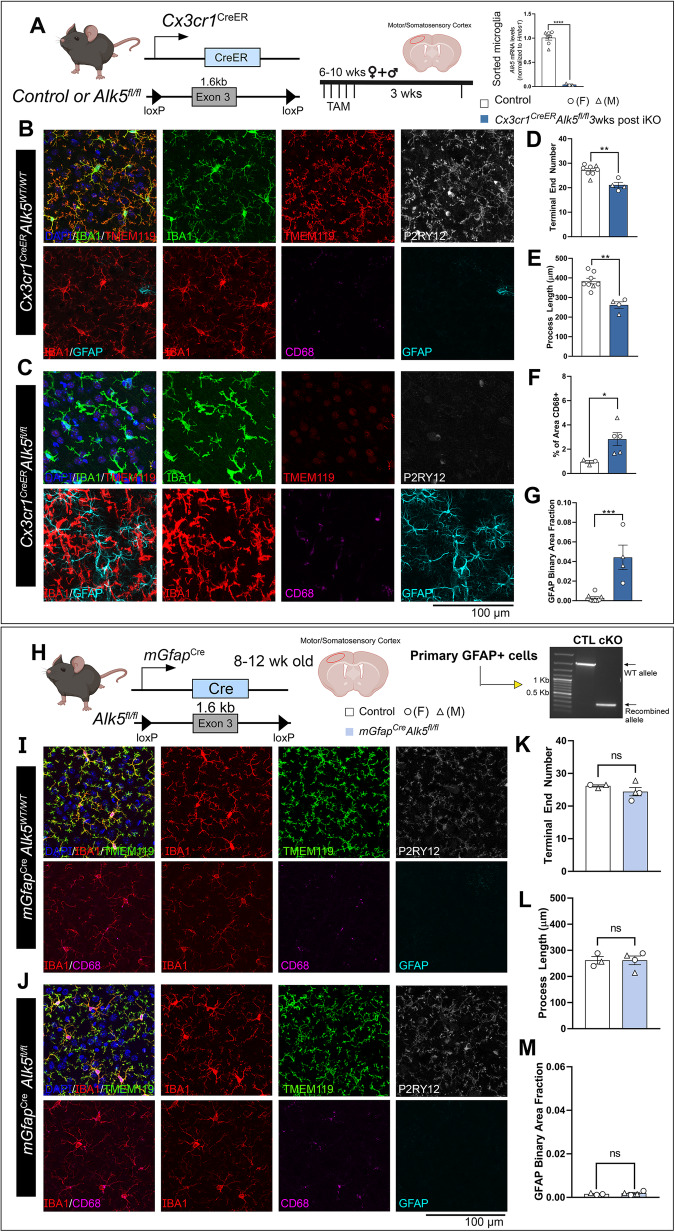
Fig. 2. ALK5-dependent TGF-β signaling is required for the maintenance of microglial homeostasis but not for astrocytic homeostasis.
A (left) Mouse model for targeting microglial TGF-β type 1 receptor Alk5 and experimental timeline. (Right) indicates the gene deletion efficiency in FACS-isolated microglia (n = 7 control, n = 3 iKO, p < 0.0001). B, C Representative immunohistochemistry images of IBA1, TMEM119, P2RY12, CD68, and GFAP in the cortex of B control animals and C Cx3cr1CreER(Jung)Alk5fl/fl knockouts 3 weeks after TAM administration. Quantification of D microglial process terminal end numbers, E total microglial process length, (D, E n = 8 control, n = 4 iKO, p = 0.001 for (D) and p = 0.0004 for (E)), F % of CD68 immunoreactive positive area (n = 3 control, n = 5 iKO, p = 0.0396), and G GFAP immunoreactive positive area fraction (n = 8 control, n = 4 iKO, p = 0.002, Mann Whitney test, two-sided). H (left) Mouse model for targeting astrocytic Alk5 and experimental timeline. (right) Indicates the gene recombination efficiency in cultured GFAP+ primary cells isolated from control or cKO mice using a PCR primer set that flanks the entire loxP-Alk5-loxP cassette. Cells from three different cKO mice show similar results. I, J Representative immunohistochemistry images of IBA1, TMEM119, P2RY12, CD68, and GFAP in the cortex of I control animals, J mGfapCreTgfb1fl/fl constitutive knockouts at 12-weeks-old. Quantification of K microglial process terminal end numbers, L total microglial process length, and M GFAP immunoreactive positive area fraction (K–M n = 3 control, n = 4 cKO). *p < 0.05, **p < 0.01, ***p < 0.001, ns = not significant. Mean ± SE, All panels are analyzed by two-sided Student’s t-test, except panel (G). (>40 Microglia were quantified for each animal and the average from one mouse was plotted as a single data point in the figure panel and treated as n = 1 for statistical analysis). Scale bar = 100 µm. A, H, Left, were created with BioRender.com and released under a creative commons attribution-noncommercial-noderivs 4.0 International license. Source data are provided as a source data file.