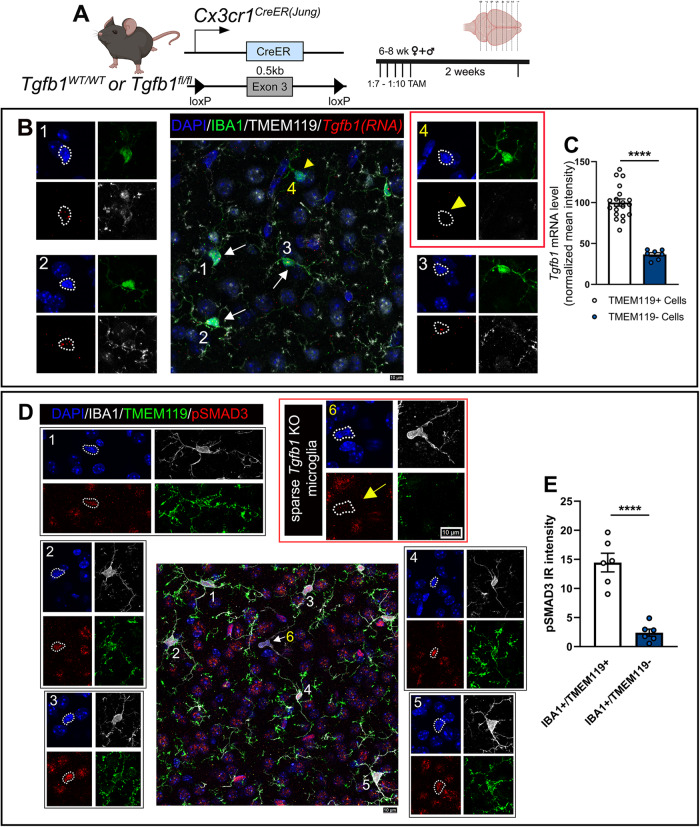
Fig. 6. In situ RNA-scope and IHC double labeling confirm loss of Tgfb1 mRNA and downregulation of TGF-β downstream signaling (pSMAD3) in dyshomeostatic individual microglia in the sparse Tgfb1 iKO model.
A Mouse model was used to examine sparse iKO in microglia and the experimental timeline with TAM dosage. B Representative image showing combined immunohistochemistry staining (for IBA1, TMEM119, and DAPI) and Tgfb1 RNA-scope hybridization. (B1–3) Surrounding normal microglia showing TMEM119 expression and Tgfb1 mRNA presence. (B4) A single microglia cell with loss of TMEM119 expression and loss of Tgfb1 mRNA. White arrows were used to mark normal cells in the central panel. Yellow arrowhead is used to mark individual iKO microglia. Note that tissue treatment for RNAscope analysis makes the IHC condition less ideal for morphology evaluation than regular IHC staining, however, IBA1 and TMEM119 expression are still distinguishable for individual WT or iKO microglia. C Quantification of RNAscope signal intensity for Tgfb1 probe in TMEM119+ and TMEM119− cells (n = 20 and 7 for control and TMEM119- group, ****p < 0.0001, Welch’s t-test, two-sided). D Representative image showing co-immunohistochemical staining with DAPI, IBA1, TMEM119, and pSMAD3. (D1–5) Surrounding normal microglia showing TMEM119 expression and pSMAD3 immunostaining. (D6) A single microglia cell with loss of TMEM119 expression and loss of pSMAD3 labeling. The yellow arrow (microglia #6) marks the individual iKO microglia. E Quantification of pSMAD3 immunoreactive intensity in TMEM119+ and TMEM119− cells (n = 6 mice for each group, ****p < 0.0001, Student’s t-test, two-sided). Scale bar = 10 µm. Mean ± SE. For additional representative images see Supplementary Fig. 12. A Created with BioRender.com and released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license. Source data are provided as a source data file.