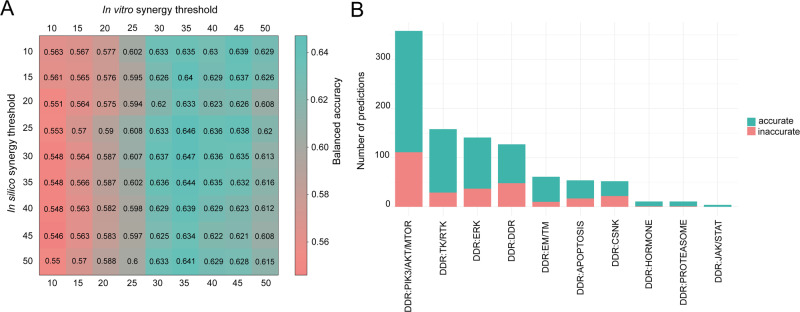
Fig. 6. Benchmarking in silico combination predictions to in vitro synergy measurements.
A In silico balanced accuracy across various synergy thresholds. Matrix represents the threshold value-dependent consensus (expressed in Balanced Accuracy, BA= (TPR + TNR) /2) between in vitro and in silico synergy. In the DREAM challenge, a threshold of 20 for both in vitro and in silico synergy was used to distinguish synergistic and non-synergistic combinations (Methods). However, the DREAM in vitro results were scaled between 0 and 100, while our in silico predictions are between 0 and 1, therefore the in silico threshold of 0.2 would correspond to it in this case. If we would use this cutoff value for the same endeavor as a gold standard, the balanced accuracy would be just under 0.6 (BA = 0.575, sensitivity=0.37, specificity=0.78). When we set the in vitro threshold for synergy to 30 and 20 for in silico synergy, our predictive power in balanced accuracy significantly improved (BA = 0.62, sensitivity=0.46, specificity=0.78). By increasing the in vitro threshold, the number of synergistic data points decreased, and the model was able to correctly detect a bigger proportion of synergistic combinations. This shows that in silico predictions are more accurate when stronger synergy is observed in vitro. B Proportion of accurate and inaccurate simulated measurements in different combination MoA groups. Due to the low in vitro data coverage, in many cases, benchmarking was not possible. Both in vitro and in silico synergy thresholds were set at 20, e.g., if both values were bigger or equal to 20, we considered that combination-cell line pair to be accurately synergistic and vice versa. Proportion of accurate and inaccurate simulations was calculated compared to the 977 overall data points. MoA groups are sorted based on the descending proportion of accurate simulations. CSNK Casein kinase, DDR DNA damage repair, EM/TM epigenetic/transcriptomic modulation, ERK extracellular signal-regulated kinase, JAK/STAT Janus kinase/signal transducers and activators of transcription, NFKB NF-kappa B, PIK3/AKT/MTOR phosphatidylinositol 3’ -kinase(PI3K)-AKT-mTOR, TK/RTK tyrosine kinase/receptor tyrosine kinase.