Summary
Comprehensive characterization of small-molecule degraders, including binary and ternary complex formation and degradation efficiency, is critical for bifunctional ligand development and understanding structure-activity relationships. Here, we present a protocol for the biochemical and cellular profiling of small-molecule degraders based on CoraFluor time-resolved fluorescence resonance energy transfer (TR-FRET) technology. We describe steps for labeling antibodies and proteins, tracer saturation binding, binary target engagement, ternary complex profiling, and off-rate determination. We then detail procedures for the quantification of endogenous and GFP fusion proteins in cell lysates.
For complete details on the use and execution of this protocol, please refer to Ichikawa et al.1
Subject areas: Cell-based Assays, High Throughput Screening, Molecular/Chemical Probes
Graphical abstract

Highlights
-
•
Comprehensive toolbox for systematically profiling proximity-inducing small molecules
-
•
Kinetic and thermodynamic analysis of binary and ternary ligand-target complexes
-
•
Quantitative characterization of ternary complex cooperativity (α) in high throughput
-
•
Facile quantification of endogenous and GFP fusion proteins in cell lysates
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Comprehensive characterization of small-molecule degraders, including binary and ternary complex formation and degradation efficiency, is critical for bifunctional ligand development and understanding structure-activity relationships. Here, we present a protocol for the biochemical and cellular profiling of small-molecule degraders based on CoraFluor time-resolved fluorescence resonance energy transfer (TR-FRET) technology. We describe steps for labeling antibodies and proteins, tracer saturation binding, binary target engagement, ternary complex profiling, and off-rate determination. We then detail procedures for the quantification of endogenous and GFP fusion proteins in cell lysates.
Before you begin
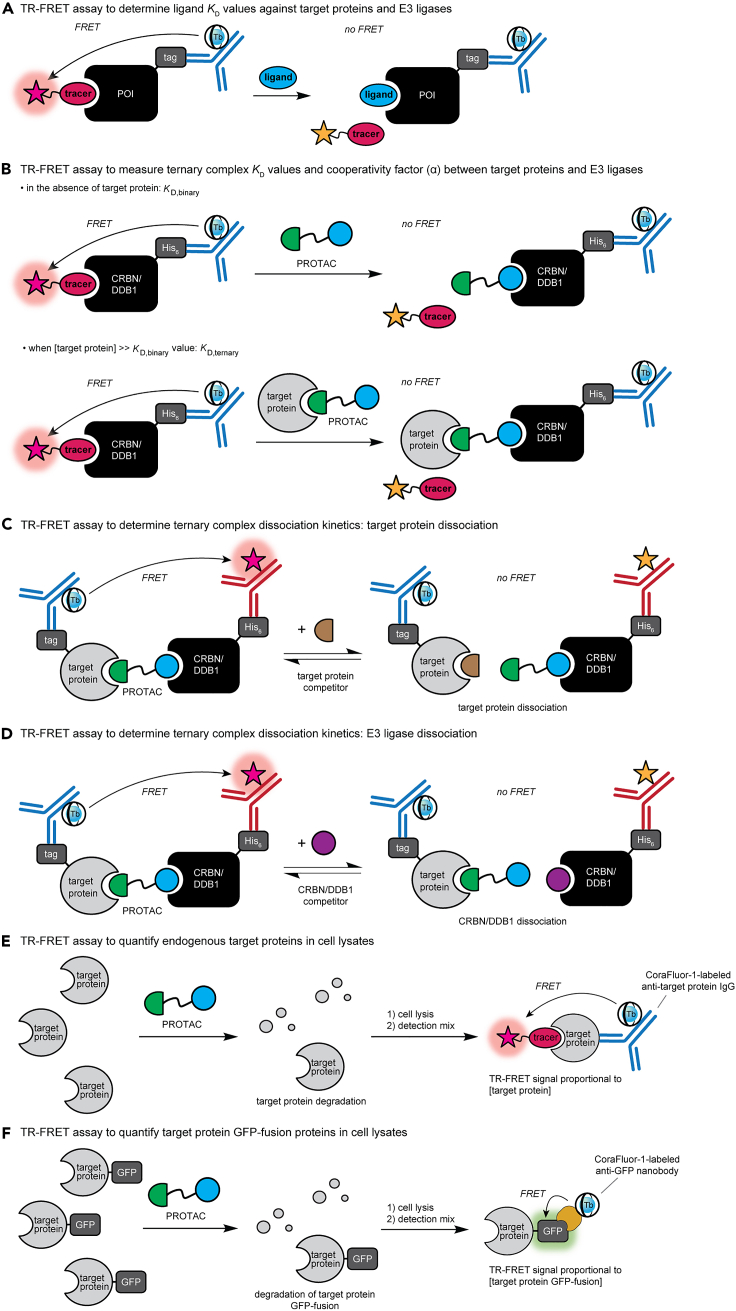
The protocols described here allow the user to determine the following parameters for hetero-bifunctional ligands and molecular glues, with a focus on small-molecule degraders, such as Cyclimids and PROTACs, that engage an E3 ligase, like cereblon (CRBN), and a desired target protein, like BRD4, resulting in degradation of the target protein. Protocols are described to measure: (i) binary target engagement (KD value) of PROTACs to His6-CRBN-DDB1 and target protein, (ii) ternary complex KD value (KD,ternary) of the target protein–PROTAC binary complex for His6-CRBN-DDB1 and direct calculation of the cooperativity factor alpha (α), (iii) measurement of the dissociation rate constant (koff) for the dissociation of the His6-CRBN-DDB1–PROTAC binary complex from the target protein, (iv) measurement of the koff value for the dissociation of the target protein–PROTAC binary complex from His6-CRBN-DDB1, (v) cellular efficacy of PROTACs of interest in degrading endogenous BRD4 or other target proteins, and (vi) cellular efficacy of PROTACs of interest in degrading target proteins expressed as GFP-fusions [here shown for BRD4(BD1)-GFP and BRD4(BD2)-GFP]. Many of these assay formats are target-agnostic and can readily be applied to other systems of interest (see Figure 1).
Figure 1.
Overview of TR-FRET assay strategies for small molecule degrader characterization
(A) TR-FRET ligand displacement assay, which is used to measure ligand affinity (KD values) to proteins of interest (POIs).
(B) TR-FRET ligand displacement assay for heterobifunctional molecules in the presence of an E3 ligase (here CRBN/DDB1) and high concentration of target protein, which allows for the measurement of the KD,ternary value for the heterobifunctional molecule-target protein binary complex toward the E3 ligase.
(C and D) TR-FRET assay to measure ternary complex dissociation kinetics, following dissociation of the target protein (C) or E3 ligase (D).
(E and F) TR-FRET assay to quantify endogenous target protein degradation (E) or target protein GFP-fusions (F) in cell lysates after compound treatment.
Preparation of CoraFluor-1 and AF488-labeled antibodies and proteins
Timing: 3 h
This protocol is for labeling antibodies or proteins of interest with CoraFluor-1-Pfp ester2 or AF488-Tfp ester for downstream TR-FRET assay development.
CRITICAL: Antibodies and proteins must be free of bovine serum albumin (BSA) or other carrier proteins, which can reduce labeling efficiency by competing with the antibody or protein of interest for dye labeling.
-
1.Buffer exchange antibody or protein into Protein Reaction Buffer using 0.5 mL, 7K MWCO Zeba spin desalting column.
-
a.Remove column bottom closure of 0.5 mL, 7K MWCO Zeba spin desalting column and loosen cap (do not completely remove the cap).
-
b.Place column in a 1.5 mL microcentrifuge tube.
-
c.Centrifuge at 1,500 × g for 1 min to remove storage solution.
-
d.Place a mark on the outside of the column where the compacted resin is slanted upward. Place column in the microcentrifuge with the mark facing outward in all subsequent centrifugation steps.
-
e.Add 300 μL of Protein Reaction Buffer on top of the resin bed, taking care to avoid disturbing the resin bed. Centrifuge at 1,500 × g for 1 min to elute buffer.
-
f.Repeat Step “e” two additional times so that the column has been equilibrated with 3 × 300 μL of Protein Reaction Buffer. The column is now equilibrated with Protein Reaction Buffer.
-
g.Place column in a new 1.5–2.0 mL microcentrifuge tube.
-
h.Slowly apply 30–130 μL of antibody or protein sample to the center of the compacted resin bed. The sample should fully enter the resin bed.
-
i.Add 10 μL of Protein Reaction Buffer to the resin bed after the sample has fully entered the resin bed. This helps with maximizing recovery.
-
j.Centrifuge at 1,500 × g for 2 min to collect sample.
-
k.This procedure should yield approximately 110 μL of antibody sample that has been buffer exchanged into reaction buffer (if starting with 100 μL sample).
-
a.
-
2.Reaction of buffer-exchanged antibody or protein with CoraFluor-1-Pfp ester or AF488-Tfp ester.Note: CoraFluor-1-Pfp may be prepared as previously described2 or purchased from Tocris (Cat. #7920). AF488-Tfp ester may be purchased from ThermoFisher (Cat. #A37570). If CoraFluor-1-Pfp is prepared as previously described2 and there is sufficient material, prepare a 2.5 mM stock solution in N,N-dimethylacetamide. If purchasing CoraFluor-1-Pfp from Tocris (Cat. #7920), add the buffer-exchanged antibody or protein solution directly to the vial containing 20 μg of CoraFluor-1-Pfp ester, such that the desired molar excess of CoraFluor-1-Pfp ester to antibody or protein is achieved. For AF488-Tfp labeling, dissolve one 100 μg aliquot of AF488-Tfp in 45.7 μL of N,N-dimethylacetamide to achieve a 2.5 mM stock solution.
-
a.Mix the desired equivalents of CoraFluor-1-Pfp ester or AF488-Tfp ester with the antibody or protein solution in Protein Reaction Buffer. Mix rapidly by vortexing and incubate for 1 h at 20°C–22°C.Note: Do not exceed 5% (v/v) organic solvent during the reaction. In our experience, a molar ratio of ∼12–15× CoraFluor-1-Pfp or AF488-Tfp to antibody yields a degree-of-labeling (DOL) of 4–6 CoraFluor-1 or AF488 labels per molecule of antibody; for proteins, a molar ratio of ∼2–3× CoraFluor-1-Pfp or AF488-Tfp to average molecular weight proteins (∼50 kDa) yields a DOL of 1–2 CoraFluor-1 or AF488 labels per molecule of protein. Equivalents may be adjusted as necessary.
-
a.
-
3.Purification of CoraFluor-1 and AF488-labeled antibody and protein conjugates.
-
a.Remove column bottom closure of 0.5 mL, 7K MWCO Zeba spin desalting column and loosen cap (do not completely remove the cap).
-
b.Place column in 1.5 mL microcentrifuge tube.
-
c.Centrifuge at 1,500 × g for 1 min to remove storage solution.
-
d.Place a mark on the outside of the column where the compacted resin is slanted upward. Place column in the microcentrifuge with the mark facing outward in all subsequent centrifugation steps.
-
e.Add 300 μL of the desired Antibody Storage Buffer on top of the resin bed, taking care to avoid disturbing the resin bed. Centrifuge at 1,500 × g for 1 min to remove buffer.Note: In our experience, for antibodies, a compatible storage buffer is 50% (v/v) phosphate-buffered saline (PBS)/glycerol, pH 7.4. Storage buffers for individual labeled antibodies and proteins may be optimized by the user.
-
f.Repeat Step “e” two additional times so that the column has been equilibrated with 3 × 300 μL of desired storage buffer. The column is now equilibrated with desired storage buffer.
-
g.Place column in a new 1.5–2.0 mL microcentrifuge tube.
-
h.Slowly apply 30–130 μL of antibody or protein reaction sample to the center of the compacted resin bed. The sample should fully enter the resin bed.
-
i.Add 10 μL of desired storage buffer to the resin bed after the sample has fully entered the resin bed. This helps with maximizing recovery.
-
j.Centrifuge at 1,500 × g for 2 min to collect sample.
-
k.This procedure should yield approximately 120 μL of labeled antibody or protein sample in desired storage buffer (if starting with 100 μL sample).
-
a.
-
4.Determination of labeled antibody or protein conjugate concentration and DOL.
-
a.For CoraFluor-1-labeled conjugates, use a NanoDrop (ThermoFisher, ND-1000) to measure absorbance at A280 and A340.
-
b.For AF488-labeled conjugates, use a NanoDrop to measure absorbance at A280 and A495.
-
c.Calculate the A280,corr value using Equation 1 below:
Where c.f. is the correction factor. For CoraFluor-1, the correction factor (A280/A340) is equal to 0.157. For AF488, the correction factor (A280/A495) is equal to 0.11.(Equation 1) -
d.Calculate the concentration of antibody or protein conjugate (cconjugate) using Equation 2 below:
Where εantibody/protein is the extinction coefficient at A280 and b is path length in centimeters.(Equation 2) -
i.The theoretical ε280 values (in M−1 cm−1) for antibodies and proteins used in this protocol are as follows: IgG = 210,000; anti-GST VHH = 28,545; anti-GFP VHH = 27,055; anti-RFP VHH = 30,035; anti-rabbit Nano-Secondary = 24,075; His6-CRBN-DDB1 = 167,000.
-
i.
-
e.Calculate the concentration of CoraFluor-1 or AF488 (cCoraFluor-1 or AF488) covalently bound to the conjugate using Equation 3 below:
Where εCoraFluor-1 or AF488 is the experimental extinction coefficient for CoraFluor-1 (equal to 22,000 M−1cm−1 at A340) or AF488 (equal to 71,000 M−1cm−1 at A495).(Equation 3) -
f.Calculate the DOL using Equation 4 below:
(Equation 4) Note: For antibodies, a typical DOL in our experience is approximately 4–6. For nanobodies, a typical DOL is approximately 1. For intermediate-sized proteins, a typical DOL is approximately 1–3.
-
a.
-
5.Storage of CoraFluor-1 and AF488-labeled conjugates.
-
a.Dilute the CoraFluor-1 or AF488-labeled conjugates to the desired glycerol concentration.
-
b.Store at desired temperature (e.g., 4°C, −20°C, or −80°C).
-
a.
Note: For labeled antibody conjugates, we recommend diluting to 50% (v/v) glycerol and storing at −80°C. For labeled protein conjugates, user-optimized storage conditions specific to the protein of interest may be used.
Determination of equilibrium dissociation constants (KD or KD,app) for fluorescent tracers by TR-FRET
Timing: 3 h
This assay is used to determine the KD value for a given fluorescent tracer specific to a given protein of interest. The KD value of the tracer is critical to obtain so that (i) optimal assay conditions can be determined and (ii) test compound KD values can be determined via the Cheng-Prusoff equation.3
Note: The following volumes assume a 8-point dose-response of the tracer in triplicate, with 30 μL assay volume per well in a white 384-well plate (e.g., Corning 3572). If more or less dead volume is needed, adjust volumes as necessary. Other plate formats (e.g., 96-well or 1536-well) or low-volume plates (e.g., Revvity 6008280) with 10 μL assay volume per well can also be used.
Note: TR-FRET measurements should be acquired with the following settings (Tecan SPARK plate reader): 340 nm excitation (50 nm bandwidth), 490 nm (terbium; 10 nM bandwidth), and 520 nm (FITC, AF488, GFP; 10 nm bandwidth) emission, 100 μs delay, 400 μs integration. The 490/10 nm and 520/10 nm emission channels are acquired with a 50% mirror and a dichroic 510 mirror, respectively, using independently optimized detector gain settings unless specified otherwise. The TR-FRET ratio is taken as the 520/490 nm intensity ratio on a per-well basis. Other instruments may be adapted to reflect these settings, and we note that specific settings for TR-FRET measurements are typically available from plate reader manufacturers.
-
6.KD determination for Thalidomide-FITC against His6-CRBN/DDB1, using CoraFluor-1-labeled anti-His6 antibody as TR-FRET donor.1
-
a.Prepare 2 mL CoraFluor-1-labeled anti-His6 antibody (1 nM final concentration) in TR-FRET Assay Buffer. Ensure complete mixing to homogeny by vortexing.
-
b.Split the 2 mL into 2 × 1 mL aliquots.
-
c.To one aliquot (POSITIVE SOLUTION), add His6-CRBN-DDB1 (2.5 nM final concentration). Ensure complete mixing to homogeny by vortexing.Note: The amount of His6-CRBN-DDB1 should be <5% of the 1 mL total volume. This ensures that there is not a substantial difference in the antibody/protein/buffer components between the POSITIVE SOLUTION and the NEGATIVE SOLUTION. An alternative would be to prepare a more concentrated mixture and adjust the volumes of each solution to 1 mL afterwards.
-
d.To the other aliquot (NEGATIVE SOLUTION), add an equal volume of water. Ensure complete mixing to homogeny by vortexing.
-
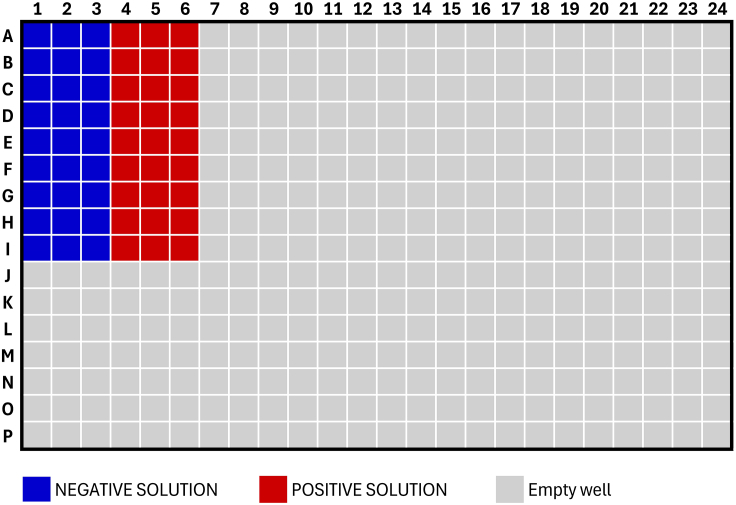
e.Add 30 μL of NEGATIVE SOLUTION and 30 μL of POSITIVE SOLUTION to wells according to the plate map in Figure 2.
-
f.Spin plate at 200 × g for 1 min to ensure all fluid is at the bottom of the well.
-
g.Dispense Thalidomide-FITC tracer by row (1–2.5 dilution step) from 100 μM stock in DMSO, e.g., using a HP D300 instrument.Note: Thalidomide-FITC cmax = 1,000 nM starting from Row A. Include 9th step (Row I) as a zero-tracer control.
-
h.Incubate plate at 20°C–22°C, covered with a lid or dummy plate to prevent evaporation.
-
i.Take TR-FRET reads at t = 10 min and t = 1 h.Note: If binding has not reached equilibrium within t = 1 h, then take TR-FRET reads at additional later timepoints (e.g., t = 2 h and t = 4 h) until the TR-FRET ratio remains unchanged. Equilibrium binding is achieved when the KD value of the tracer stabilizes to a consistent value.
-
j.Determine the Corrected TR-FRET ratios by subtracting the average TR-FRET ratio of the three NEGATIVE SOLUTION replicates from the TR-FRET ratios of the three POSITIVE SOLUTION replicates on a per-row basis.
-
k.Plot the Corrected TR-FRET ratios against the linear concentration of tracer. Fit the data to One Site – Specific Binding model as shown in Equation 5 below:
(Equation 5) Note:Bmax is the maximum specific binding in the same units as Y. -
l.Adjust the KD value for the one-site model to account for a two-site model due to the presence of the bivalent anti-His6 antibody, as shown in Equation 6 below:
(Equation 6) Note: The KD,2-site value for Thalidomide-FITC should be approximately 117 nM.
-
a.
-
7.KD determination for Thalidomide-FITC against CoraFluor-1-labeled His6-CRBN-DDB1.
-
a.Prepare 2 mL CoraFluor-1-labeled His6-CRBN-DDB1 (2.5 nM final concentration) in TR-FRET Assay Buffer. Ensure complete mixing to homogeny by vortexing.
-
b.Split the 2 mL into 2 × 1 mL aliquots.
-
c.To one aliquot (POSITIVE SOLUTION), add DMSO (1% (v/v) final concentration). Ensure complete mixing to homogeny by vortexing.
-
d.To the other aliquot (NEGATIVE SOLUTION), add 100 μM Lenalidomide-5′-NH2 (from 10 mM stock solution in DMSO, 1% (v/v) final DMSO concentration). Ensure complete mixing to homogeny by vortexing.
-
e.Add 30 μL of NEGATIVE SOLUTION and 30 μL of POSITIVE SOLUTION to wells according to plate map in Figure 2.
-
f.Spin plate at 200 × g for 1 min to ensure all fluid is at the bottom of the well.
-
g.Dispense Thalidomide-FITC tracer by row (1–2.5 dilution step) from 100 μM stock in DMSO, e.g., using HP D300.Note: Thalidomide-FITC cmax = 1,000 nM starting from Row A. Include 9th step (Row I) as zero-tracer control.
-
h.Incubate plate at 20°C–22°C, covered with a lid or dummy plate to prevent evaporation.
-
i.Take TR-FRET reads at t = 10 min and t = 1 h.Note: If binding has not reached equilibrium within t = 1 h, then take TR-FRET reads at t = 2 h and t = 4 h.
-
j.Determine the Corrected TR-FRET Ratios by subtracting the average TR-FRET ratio of the three NEGATIVE SOLUTION replicates from the TR-FRET ratios of the three POSITIVE SOLUTION replicates on a per-row basis.
-
k.Plot the Corrected TR-FRET Ratios against the linear concentration of tracer. Fit the data to One Site – Specific Binding model as shown in Equation 5.Note: The KD,1-site value for Thalidomide-FITC should be approximately 301 nM. This KD,1-site value was obtained using CoraFluor-1-labeled His6-CRBN-DDB1 with a DOL of 4.6. Because the protein is being labeled directly with CoraFluor-1-Pfp ester in these experiments, the KD,1-site of Thalidomide-FITC and test compounds may be dependent on the DOL of the construct.
-
a.
-
8.KD determination for JQ1-FITC against GST-BRD4(BD1) or GST-BRD4(BD2), using CoraFluor-1-labeled anti-GST VHH as TR-FRET donor.4,5
-
a.Prepare 2 mL CoraFluor-1-labeled anti-GST-VHH (4 nM final concentration) in TR-FRET Assay Buffer. Ensure complete mixing to homogeny by vortexing.
-
b.Split the 2 mL into 2 × 1 mL aliquots.
-
c.To one aliquot (POSITIVE SOLUTION), add GST-BRD4(BD1) or GST-BRD4(BD2) (0.5 nM final concentration). Ensure complete mixing to homogeny by vortexing.Note: The amount of GST-BRD4(BD1) or GST-BRD4(BD2) should be <5% of the 1 mL total volume.
-
d.To the other aliquot (NEGATIVE SOLUTION), add an equal volume of water. Ensure complete mixing to homogeny by vortexing.
-
e.Add 30 μL of NEGATIVE SOLUTION and 30 μL of POSITIVE SOLUTION to wells according to plate map in Figure 2.
-
f.Spin plate at 200 × g for 1 min to ensure all fluid is at the bottom of the well.
-
g.Dispense JQ1-FITC tracer by row (1–2.5 dilution step) from 10 μM stock in DMSO, e.g., using a HP D300 instrument.Note: JQ1-FITC cmax = 100 nM starting from Row A. Include 9th step (Row I) as zero-tracer control.
-
h.Incubate plate at 20°C–22°C, covered with a lid or dummy plate to prevent evaporation.
-
i.Take TR-FRET reads at t = 10 min and t = 1 h.Note: If binding has not reached equilibrium within t = 1 h, then take TR-FRET reads at t = 2 h and t = 4 h.
-
j.Determine the Corrected TR-FRET Ratios by subtracting the average TR-FRET ratio of the three NEGATIVE SOLUTION replicates from the TR-FRET ratios of the three POSITIVE SOLUTION replicates on a per-row basis.
-
k.Plot the Corrected TR-FRET Ratios against the linear concentration of tracer. Fit the data to One Site – Specific Binding model as shown in Equation 5.
-
l.Adjust the KD value for the one-site model to account for a two-site model due to the presence of the dimeric GST protein as shown in Equation 6.Note: The KD,2-site values for JQ1-FITC should be approximately 16 nM and 14 nM for GST-BRD4(BD1) and GST-BRD4(BD1), respectively.
-
a.
-
9.KD determination for SLF-FITC against His6-FKBP12, using CoraFluor-1-labeled anti-His6 antibody as TR-FRET donor.1
-
a.Prepare 2 mL CoraFluor-1-labeled anti-His6 antibody (2.5 nM final concentration) in TR-FRET Assay Buffer. Ensure complete mixing to homogeny by vortexing.
-
b.Split the 2 mL into 2 × 1 mL aliquots.
-
c.To one aliquot (POSITIVE SOLUTION), add His6-FKBP12 (5 nM final concentration). Ensure complete mixing to homogeny by vortexing.Note: The amount of His6-FKBP12 should be <5% of the 1 mL total volume.
-
d.To the other aliquot (NEGATIVE SOLUTION), add an equal volume of water. Ensure complete mixing to homogeny by vortexing.
-
e.Add 30 μL of NEGATIVE SOLUTION and 30 μL of POSITIVE SOLUTION to wells according to plate map in Figure 2.
-
f.Spin plate at 200 × g for 1 min to ensure all fluid is at the bottom of the well.
-
g.Dispense SLF-FITC tracer by row (1–2.5 dilution step) from 100 μM stock in DMSO, e.g., using HP D300.Note: SLF-FITC cmax = 1,000 nM starting from Row A. Include 9th step (Row I) as zero-tracer control.
-
h.Incubate plate at 20°C–22°C, covered with a lid or dummy plate to prevent evaporation.
-
i.Take TR-FRET reads at t = 10 min and t = 1 h.Note: If binding has not reached equilibrium within t = 1 h, then take TR-FRET reads at t = 2 h and t = 4 h.
-
j.Determine the Corrected TR-FRET Ratios by subtracting the average TR-FRET ratio of the three NEGATIVE SOLUTION replicates from the TR-FRET ratios of the three POSITIVE SOLUTION replicates on a per-row basis.
-
k.Plot the Corrected TR-FRET Ratios against the linear concentration of tracer. Fit the data to One-Site – Specific Binding model as shown in Equation 5.
-
l.Adjust the KD value for the one-site model to account for a two-site model due to the presence of the bivalent anti-His6 antibody, as shown in Equation 6.Note: The KD,2-site value for SLF-FITC should be approximately 87 nM.
-
a.
-
10.KD determination for Palbociclib-FITC against His6-GST-CDK4/GST-CD3 or His6-CDK6/GST-CD1, using CoraFluor-1-labeled anti-His6 antibody as TR-FRET donor.
-
a.Prepare 2 mL CoraFluor-1-labeled anti-His6 antibody (1 nM final concentration) in TR-FRET Assay Buffer. Ensure complete mixing to homogeny by vortexing.
-
b.Split the 2 mL into 2 × 1 mL aliquots.
-
c.To one aliquot (POSITIVE SOLUTION), add His6-GST-CDK4/GST-CD3 or His6-CDK6/GST-CD1 (0.5 nM or 2 nM final concentration, respectively). Ensure complete mixing to homogeny by vortexing.Note: The amount of His6-GST-CDK4/GST-CD3 or His6-CDK6/GST-CD1 should be <5% of the 1 mL total volume.
-
d.To the other aliquot (NEGATIVE SOLUTION), add an equal volume of water. Ensure complete mixing to homogeny by vortexing.
-
e.Add 30 μL of NEGATIVE SOLUTION and 30 μL of POSITIVE SOLUTION to wells according to plate map in Figure 2.
-
f.Spin plate at 200 × g for 1 min to ensure all fluid is at the bottom of the well.
-
g.Dispense Palbociclib-FITC tracer by row (1–2.5 dilution step) from 100 μM stock in DMSO, e.g., using HP D300.Note: Palbociclib-FITC cmax = 1,000 nM starting from Row A. Include 9th step (Row I) as zero-tracer control.
-
h.Incubate plate at 20°C–22°C, covered with a lid or dummy plate to prevent evaporation.
-
i.Take TR-FRET reads at t = 10 min and t = 1 h.Note: If binding has not reached equilibrium within t = 1 h, then take TR-FRET reads at t = 2 h and t = 4 h.
-
j.Determine the Corrected TR-FRET Ratios by subtracting the average TR-FRET ratio of the three NEGATIVE SOLUTION replicates from the TR-FRET ratios of the three POSITIVE SOLUTION replicates on a per-row basis.
-
k.Plot the Corrected TR-FRET Ratios against the linear concentration of tracer. Fit the data to One Site – Specific Binding model as shown in Equation 5.Note: For His6-GST-CDK4/GST-CD3 and His6-CDK6/GST-CD1, only KD,app values can be determined due to a multivalent binding model. The KD,app values for Palbociclib-FITC should be approximately 5 nM and 292 nM for His6-GST-CDK4/GST-CD3 and His6-CDK6/GST-CD1, respectively.
-
a.
Figure 2.
Plate layout for positive and negative assay solutions during saturation binding experiments
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BRD4 (E2A7×) | Cell Signaling Technology | Cat# 13440S; RRID: AB_2687578 |
| Anti-His6 | Abcam | Cat# ab18184; RRID: AB_444306 |
| Anti-GST | Abcam | Cat# ab19256; RRID: AB_444809 |
| Anti-rabbit Nano-Secondary (CTK0101) | ChromoTek | Cat# shurbGNHS-1; RRID: AB_2864267 |
| Anti-GST VHH | ChromoTek | Cat# ST-250; RRID: AB_2631368 |
| Anti-GFP VHH | ChromoTek | Cat# GT-250; RRID: AB_2631361 |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco’s modified Eagle’s medium (DMEM) | Genesee Scientific | Cat# 25-500 |
| Trypsin-EDTA | Fisher Scientific | Cat# 25200114 |
| Fetal bovine serum (FBS) | Peak Serum | Cat# PS-FB2 |
| Penicillin-streptomycin (100×) | Lonza | Cat# 17-602E |
| CoraFluor-1-Pfp | Payne et al.2 | N/A |
| CoraFluor-1-Pfp | Tocris | Cat# 7920 |
| AF488-Tfp | Thermo Fisher Scientific | Cat# A37570 |
| His6-CRBN/DDB1 | Bristol Myers Squibb Also see Lin, W. et al. for compatible construct6 |
A generous gift |
| Thal-FITC | Ichikawa et al.1 | N/A |
| GST-BRD4(BD1) | BPS | Cat# 31040 |
| GST-BRD4(BD2) | EpiCypher | Cat# 15-0013 |
| JQ1-FITC | Payne et al.4 | N/A |
| JQ1-FITC | Tocris | Cat# 7722 |
| His6-FKBP12 | Ichikawa et al.1 | N/A |
| SLF-FITC | Ichikawa et al.1 | N/A |
| His6-GST-CDK4/GST-CD3 | BPS | Cat# 40104 |
| His6-CDK6/GST-CD1 | BPS | Cat# 40097 |
| Palbociclib-FITC | Ichikawa et al.1 | N/A |
| AEBSF hydrochloride | Combi Blocks | Cat# SS-7834 |
| Benzonase nuclease | MilliporeSigma | Cat# E1014 |
| DTT | Thermo Fisher Scientific | Cat# R0861 |
| Lenalidomide-5′-NH2 | Muller et al.7 | N/A |
| Experimental models: Cell lines | ||
| MDA-MB-231 | ATCC | Cat# HTB-26 |
| HEK293T cells stably expressing stably expressing BRD4(BD1)-GFP | Nowak et al.8 | N/A |
| HEK293T cells stably expressing stably expressing BRD4(BD2)-GFP | Nowak et al.8 | N/A |
| Software and algorithms | ||
| Microsoft Excel (v16.44) | Microsoft | N/A |
| GraphPad Prism 10 | GraphPad Software | N/A |
| SparkControl V2.1 | Tecan Group Ltd. | N/A |
| Other | ||
| Spark multimode microplate reader (monochromator) | Tecan Group Ltd. | N/A |
| White 384-well plates for TR-FRET | Corning | Cat# 3572 |
| White 384-well plates for TR-FRET | Greiner | Cat# 781207 |
| White low-volume 384-well plates for TR-FRET | Revvity | Cat# 6008280 |
| 96-well plates for cell culture | Corning | Cat# 3904 |
| Jitterbug incubated microplate shaker | Boekel Scientific | Cat# 130000 |
| 8-channel, 15–1,250 μL Matrix Equalizer electronic multichannel pipette | Thermo Fisher Scientific | Cat# 2034 |
| 12-channel, 2–125 μL Matrix Equalizer electronic multichannel pipette | Thermo Fisher Scientific | Cat# 2231 |
| Multidrop Combi reagent dispenser | Thermo Fisher Scientific | Cat# 5840300 |
| 7K MWCO Zeba spin desalting columns, 0.5 mL | Thermo Fisher Scientific | Cat# 89883 |
Materials and equipment
Protein Reaction Buffer (100 mM sodium carbonate, pH 8.5, and 0.05% (v/v) TWEEN-20)
| Reagent | Final concentration | Amount |
|---|---|---|
| Sodium carbonate | 100 mM | 5.30 g |
| 20% (v/v) aqueous TWEEN-20 solution | 0.05% (v/v) | 1.25 mL |
| MilliQ water | N/A | 500 mL |
Store at 25°C for up to 3 months.
-
•
Dissolve 5.30 g sodium carbonate in 450 mL MilliQ water.
-
•
Add 1.25 mL of 20% (vol/vol) aqueous TWEEN-20 solution.
-
•
Adjust to pH 8.5 with 12 M HCl.
-
•
Add MilliQ water to final volume of 500 mL.
Antibody Storage Buffer (50 mM sodium phosphate, pH 7.4, 150 mM NaCl, 0.05% (v/v) TWEEN-20)
| Reagent | Final concentration | Amount |
|---|---|---|
| 1.0 M monosodium phosphate | 9.7 mM | 4.8 mL |
| 1.0 M disodium phosphate | 40.3 mM | 20.2 mL |
| 5.0 M NaCl | 150 mM | 15 mL |
| 20% (v/v) aqueous TWEEN-20 solution | 0.05% (v/v) | 1.25 mL |
| MilliQ water | N/A | 500 mL |
Store at 4°C for up to 3 months.
-
•
Add all buffer components to 450 mL MilliQ water.
-
•
Adjust to pH 7.4 with 12 M HCl or 5 M NaOH.
-
•
Add MilliQ water to final volume of 500 mL.
TR-FRET Assay Buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mg/mL BSA, 0.005% (v/v) TWEEN-20)
| Reagent | Final concentration | Amount |
|---|---|---|
| 500 mM HEPES | 25 mM | 25 mL |
| 5.0 M NaCl | 150 mM | 15 mL |
| 50 mg/mL BSA | 0.5 mg/mL | 5 mL |
| 20% (v/v) aqueous TWEEN-20 solution | 0.005% (v/v) | 0.125 mL |
| MilliQ water | N/A | 500 mL |
Store at 4°C for up to 3 months.
-
•
Add all buffer components to 450 mL MilliQ water.
-
•
Adjust to pH 7.5 with 12 M HCl or 5 M NaOH.
-
•
Add MilliQ water to final volume of 500 mL.
Incomplete Lysis Buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 0.2% (v/v) Triton X-100, 0.02% (v/v) TWEEN-20)
| Reagent | Final concentration | Amount |
|---|---|---|
| 500 mM HEPES | 25 mM | 25 mL |
| 5.0 M NaCl | 150 mM | 15 mL |
| 20% (v/v) aqueous Triton X-100 solution | 0.2% (v/v) | 5 mL |
| 20% (v/v) aqueous TWEEN-20 solution | 0.02% (v/v) | 0.5 mL |
| MilliQ water | N/A | 500 mL |
Store at 4°C for up to 3 months.
-
•
Add all buffer components to 450 mL MilliQ water.
-
•
Adjust to pH 7.5 with 12 M HCl or 5 M NaOH.
-
•
Add MilliQ water to final volume of 500 mL.
Complete Lysis Buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 0.2% (v/v) Triton X-100, 0.02% (v/v) TWEEN-20, 1 mM AEBSF hydrochloride, 250 U/mL Benzonase, 2 mM DTT)
| Reagent | Final concentration | Amount |
|---|---|---|
| 100 mM aqueous AEBSF hydrochloride | 1 mM | 100 μL |
| 250 U/μL Benzonase | 250 U/mL | 10 μL |
| 100 mM DTT | 2 mM | 200 μL |
| Incomplete Lysis Buffer | N/A | 9.7 mL |
Store at 4°C for up to 1 day.
-
•
Add all buffer components to 9.7 mL Incomplete Lysis Buffer.
Dilution Buffer (25 mM HEPES, pH 7.5, 150 mM NaCl, 0.005% (v/v) TWEEN-20)
| Reagent | Final concentration | Amount |
|---|---|---|
| 500 mM HEPES | 25 mM | 25 mL |
| 5.0 M NaCl | 150 mM | 15 mL |
| 20% (v/v) aqueous TWEEN-20 solution | 0.005% (v/v) | 0.125 mL |
| MilliQ water | N/A | 500 mL |
Store at 4°C for up to 3 months.
-
•
Add all buffer components to 450 mL MilliQ water.
-
•
Adjust to pH 7.5 with 12 M HCl or 5 M NaOH.
-
•
Add MilliQ water to final volume of 500 mL.
Step-by-step method details
Determining KD values of test compounds against His6-CRBN-DDB1, GST-BRD4(BD1), GST-BRD4(BD2), His6-FKBP12, His6-GST-CDK4/GST-CD3, or His6-CDK6/GST-CD1
Timing: 2 h
This assay is used to determine the affinity (KD value) of test compounds against target proteins, in this case, His6-CRBN-DDB1, GST-BRD4(BD1), GST-BRD4(BD2), His6-FKBP12, His6-GST-CDK4/GST-CD3, or His6-CDK6/GST-CD1. The assay concept is target-agnostic and may also be applied to other systems beyond those shown below.
Note: The following volumes assume a 15-point dose-response of the test compound in triplicate, with 30 μL assay volume per well in a white 384-well plate (e.g., Corning 3572). If more or less dead volume is needed, adjust volumes as necessary. Other plate formats (e.g., 96-well or 1536-well) or low-volume plates (e.g., Revvity 6008280) with 10 μL assay volume per well can also be used.
-
1.Prepare master assay solutions at 1× concentration in TR-FRET Assay Buffer.Note: The tracer is generally employed at a concentration equal to 0.5–3 × KD, which provides a good balance of sensitivity and assay performance.9 For tracers with slow off-rates, test compounds can be added to the protein solution in the well before the tracer is added, allowing the assay system to reach equilibrium faster. This approach may be relevant for tracers with t1/2 values > 25 min, which require > 2 h to equilibrate (5 × t1/2) when the tracer is added before the test compound.
-
a.To measure affinity for His6-CRBN-DDB1, use 2.5 nM His6-CRBN-DDB1, 1 nM CoraFluor-1-labeled anti-His6 antibody, and 50 nM Thalidomide-FITC.
-
b.To measure affinity for CoraFluor-1-labeled His6-CRBN-DDB1, use 2.5 nM CoraFluor-1-labeled His6-CRBN-DDB1 and 200 nM Thalidomide-FITC.
-
c.To measure affinity for GST-BRD4(BD1), use 4 nM GST-BRD4(BD1), 4 nM CoraFluor-1-labeled anti-GST VHH, and 20 nM JQ1-FITC.
-
d.To measure affinity for GST-BRD4(BD2), use 4 nM GST-BRD4(BD2), 4 nM CoraFluor-1-labeled anti-GST VHH, and 20 nM JQ1-FITC.
-
e.To measure affinity for His6-FKBP12, use 5 nM His6-FKBP12, 2.5 nM CoraFluor-1-labeled anti-His6 antibody, and 60 nM SLF-FITC.
-
f.To measure affinity for His6-GST-CDK4/GST-CD3, use 2 nM His6-GST-CDK4/GST-CD3, 1 nM CoraFluor-1-labeled anti-His6 antibody, and 30 nM Palbociclib-FITC.
-
g.To measure affinity for His6-CDK6/GST-CD1, use 2 nM His6-CDK6/GST-CD1, 1 nM CoraFluor-1-labeled anti-His6 antibody, and 200 nM Palbociclib-FITC.Note: Tracer concentration may need to be optimized for different plate readers.
-
a.
-
2.
Dispense 30 μL of assay solution to all wells of the 384-well plate using a Multidrop Combi dispenser.
-
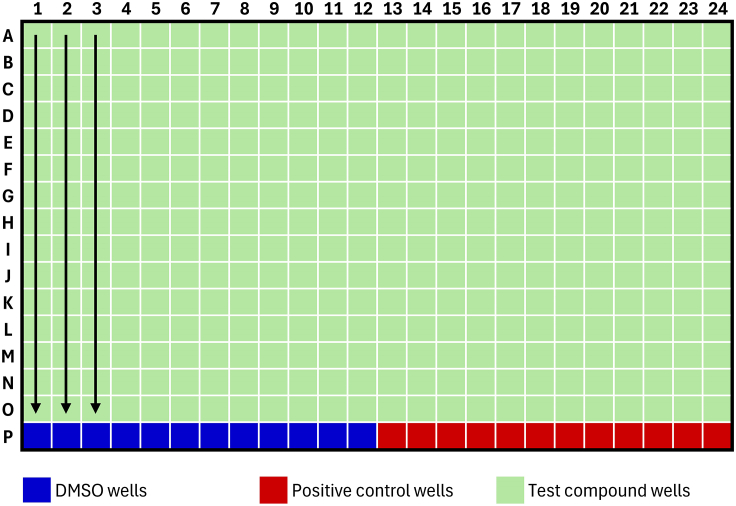
3.Dispense compounds (e.g., via HP D300) according to plate map in Figure 3. The standard dose-response range is from 10 μM to 0.6 nM (15-point dose-response, 2-fold dilution steps).
-
4.
Normalize all wells to the same DMSO content [usually 1% (v/v)].
-
5.
Incubate for 2 h or until assay has reached equilibrium (5 × t1/2 of tracer).
-
6.
Take the TR-FRET ratio on a per-well basis.
-
7.
Calculate the Normalized TR-FRET Ratios on a per-well basis, using Equation 7 below:
| (Equation 7) |
-
8.
Plot the Normalized TR-FRET Ratios against the log10 concentration of test compound. Fit the data to a log(inhibitor) vs. response – variable slope (four parameter) dose-response model as shown in Equation 8 below:
| (Equation 8) |
-
9.
Calculate the KD or KD,app of the test compound, using Equation 9 below:
| (Equation 9) |
Where IC50 is the IC50 of the test compound, [S] is the concentration of tracer used in the assay, and KX is the KD or KD,app of the tracer used in the assay.
Figure 3.
Standard plate map for TR-FRET ligand displacement assay experiments
Arrows indicate decreasing compound concentration for one compound in triplicate. In this format, 8 compounds can be fitted per 384-well plate.
Determining ternary complex KD values (KD,ternary) and cooperativity factor alpha (α) values of test compounds with His6-CRBN-DDB1 and target proteins
Timing: 2 h
This assay is used to determine the ternary complex affinity (KD,ternary value) and cooperativity factor alpha (α) of test compounds between His6-CRBN-DDB1 and target proteins, in this case, GST-BRD4(BD1), GST-BRD4(BD2), and His6-FKBP12. The assay concept is nearly identical to a standard TR-FRET ligand displacement assay, however in this case the target protein is added to the assay mix at a high concentration (≥10-fold relative to KD of PROTAC to target protein) that results in the majority (≥90%) of the PROTAC of interest being saturated with target protein.9 Therefore, what is effectively being titrated against His6-CRBN-DDB1 is the target protein–PROTAC binary complex, which greatly simplifies the determination of KD,ternary and α values.8,13,14,15,16,17 While the example shown here is for His6-CRBN-DDB1 and target proteins of interest, the assay concept is target-agnostic and may be extended to other systems.
CRITICAL: When using CRBN-DDB1 with a His6-tag to install the TR-FRET donor (CoraFluor-1-labeled anti-His6 antibody in this case), it is critical that the target protein does not also contain a His6-tag, which will interfere with the assay. Therefore, orthogonal tags on the target protein must be used (e.g., GST, FLAG), or this His6-tag on the target protein must be cleaved off before the assay is performed. Alternatively, as is shown in the example with His6-FKBP12, His6-CRBN-DDB1 may be directly labeled with CoraFluor-1-Pfp ester, which eliminates the need to use the CoraFluor-1-labeled anti-His6 antibody and is therefore compatible with target proteins containing a His6-tag. Finally, it is important to use the KD,binary values from the same assay system (e.g., His6-CRBN-DDB1 + CoraFluor-1-labeled anti-His6 antibody versus CoraFluor-1-labeled His6-CRBN-DDB1) when calculating the cooperativity factor α.
Note: The following volumes assume an 8-point dose-response of the tracer in duplicate, with 10 μL assay volume per well in a white 384-well plate (e.g., Revvity 6008280).
-
10.Prepare master assay solutions at 1× concentration in TR-FRET Assay Buffer.
-
a.For the system consisting of His6-CRBN-DDB1, CoraFluor-1-labeled anti-His6 antibody, Thalidomide-FITC, and GST-BRD4(BD1) or GST-BRD4(BD2), use 2.5 nM His6-CRBN-DDB1, 1 nM CoraFluor-1-labeled anti-His6 antibody, 50 nM Thalidomide-FITC, and 1 μM GST-BRD4(BD1) or GST-BRD4(BD2).Note: Concentration of BRD4 or any target protein should be optimized individually, as discussed above. The concentration of target protein used should be ≥10-fold relative to the KD of the PROTAC to the target protein. Tracer concentration may need to be optimized for different plate readers.
-
b.For the system consisting of CoraFluor-1-labeled His6-CRBN-DDB1, Thalidomide-FITC, and His6-FKBP12, use 2.5 nM CoraFluor-1-labeled His6-CRBN-DDB1, 200 nM Thalidomide-FITC, and 1 μM His6-FKBP12.
-
a.
-
11.
Dispense 10 μL of assay solution to all wells of the 384-well plate using a Multidrop Combi dispenser or via manual pipetting.
-
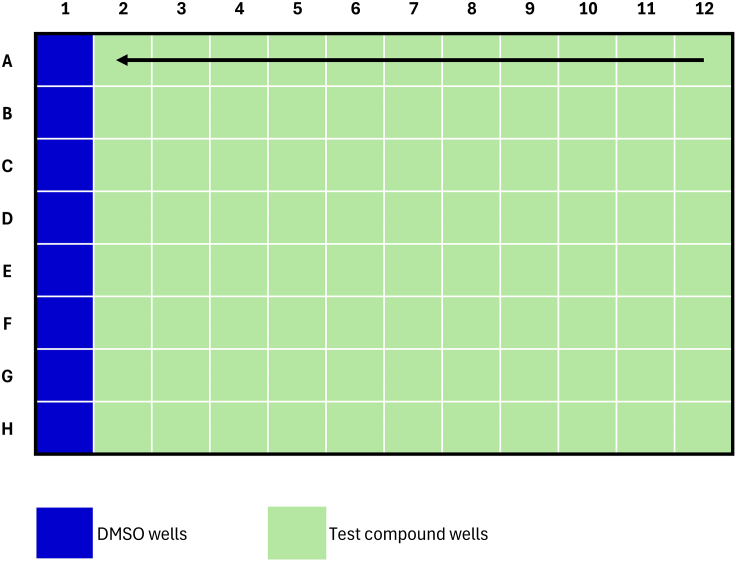
12.Dispense compounds (e.g., via HP D300) according to plate map in Figure 4. The standard dose-response range is from 1 μM to 0.06 nM (8-point dose-response, 4-fold dilution steps).
-
a.For assays with His6-CRBN-DDB1, dBET6 can be used as a positive control.
-
a.
-
13.
Normalize all wells to the same DMSO content [usually 1% (v/v)].
-
14.
Incubate for 2 h or until assay has reached equilibrium (5 × t1/2 of tracer).
-
15.
Take the TR-FRET ratio on a per-well basis.
-
16.
Calculate the Normalized TR-FRET Ratios on a per-well basis using Equation 7.
-
17.
Plot the Normalized TR-FRET Ratios against the log10 concentration of test compound. Fit the data to a log(inhibitor) vs. response – variable slope (four parameter) dose-response model using Equation 8.
-
18.
Calculate the KD,ternary using Equation 9.
-
19.
Calculate α using Equation 10 below:
| (Equation 10) |
Where KD,binary and KD,ternary are the respective KD values for the PROTACs for His6-CRBN-DDB1 in the absence and presence of target protein, respectively.
Figure 4.
Standard plate map for TR-FRET KD,ternary and α assay experiments
Arrows indicate decreasing compound concentration for one compound in duplicate. In this format, 23 compounds can be fitted per 384-well plate.
Determining dissociation rate constant (koff) values for PROTAC-mediated ternary complexes between His6-CRBN-DDB1 and target proteins
Timing: 2 h
This assay is used to either measure (i) dissociation of the His6-CRBN-DDB1–PROTAC binary complex from the target protein, or (ii) dissociation of the target protein–PROTAC binary complex from His6-CRBN-DDB1. Following addition of a competing ligand at the target protein site (e.g., JQ1 for BRD4), re-binding of the His6-CRBN-DDB1–PROTAC binary complex is prevented, which allows for measurement (i) above. In contrast, following addition of a competing ligand at the His6-CRBN-DDB1 site (e.g., Lenalidomide-5′-NH2), re-binding of the target protein–PROTAC binary complex is prevented, which allows for measurement (ii) above. As before, while the example shown here is for His6-CRBN-DDB1 and target proteins of interest, the assay concept is target-agnostic and may be extended to other systems.
CRITICAL: As before, when using CRBN-DDB1 with a His6-tag to install the TR-FRET donor (CoraFluor-1-labeled anti-His6 antibody in this case), it is critical that the target protein does not also contain a His6-tag, which will interfere with the assay. Therefore, orthogonal tags on the target protein must be used (e.g., GST, FLAG), or the His6-tag on the target protein must be cleaved off before the assay is performed.
Note: The following volumes assume duplicate wells, with 10 μL assay volume per well in a white 384-well plate (e.g., Revvity 6008280).
-
20.Prepare master assay solutions at 1× concentration in TR-FRET Assay Buffer.
 CRITICAL: When performing these assays, in our experience 10 nM of each protein (His6-CRBN-DDB1 and target protein) is sufficient for robust signal. Under standard conditions, the PROTAC is added at a concentration equal to its KD,ternary value, specific to His6-CRBN-DDB1 and the target protein. However, the concentration of PROTAC used may be optimized by the user, and may depend on (i) the KD,ternary value relative to the amount of protein used (e.g., if KD,ternary << [protein] or [E3 ligase], then the amount of ternary complex formed will be relatively small), (ii) titration effects arising from large differences in the KD,binary values of the PROTAC for the target protein and E3 ligase, (iii) the presence of multi-site binding models, which can influence the concentrations of assay components that give optimal signal. In general, the ternary complex is allowed to pre-form for 1 h at room temperature, however, for some systems, more or less time may be required to reach equilibrium before starting the off-rate assay.
CRITICAL: When performing these assays, in our experience 10 nM of each protein (His6-CRBN-DDB1 and target protein) is sufficient for robust signal. Under standard conditions, the PROTAC is added at a concentration equal to its KD,ternary value, specific to His6-CRBN-DDB1 and the target protein. However, the concentration of PROTAC used may be optimized by the user, and may depend on (i) the KD,ternary value relative to the amount of protein used (e.g., if KD,ternary << [protein] or [E3 ligase], then the amount of ternary complex formed will be relatively small), (ii) titration effects arising from large differences in the KD,binary values of the PROTAC for the target protein and E3 ligase, (iii) the presence of multi-site binding models, which can influence the concentrations of assay components that give optimal signal. In general, the ternary complex is allowed to pre-form for 1 h at room temperature, however, for some systems, more or less time may be required to reach equilibrium before starting the off-rate assay.-
a.For the system consisting of His6-CRBN-DDB1, AF488-labeled anti-His6 antibody, GST-BRD4(BD1) or GST-BRD4(BD2), and CoraFluor-1-labeled anti-GST antibody, use 10 nM His6-CRBN-DDB1, 5 nM AF488-labeled anti-His6 antibody, 10 nM GST-BRD4(BD1) or GST-BRD4(BD2), and 5 nM CoraFluor-1-labeled anti-GST antibody. Add PROTAC at a concentration equal to its KD,ternary value. For background determination, also make up assay solution containing an equal volume of DMSO in place of PROTAC.
-
a.
-
21.
Dispense 10 μL of assay solution (duplicate wells) to desired wells of the 384-well plate via manual pipetting.
-
22.
Incubate plate for 1–2 h.
-
23.
Take an initial t = 0 TR-FRET measurement to establish the assay plateau value.
-
24.
Add a concentration of competing ligand that will result in complete occupancy of the desired target site (e.g., 50 μM Lenalidomide-5′-NH2 for His6-CRBN-DDB1 or 10 μM JQ1 for BRD4), via HP D300 or manual pipetting.
-
25.
Immediately measure the time-dependent change in TR-FRET signal over the desired time frame, ideally in 10–30 s intervals, or until the TR-FRET signal of PROTAC-containing wells equals that of DMSO-containing wells (background).
Note: Plate readers equipped with injectors can be helpful to capture fast-off kinetics.
-
26.
Take the TR-FRET ratio on a per-well basis as a function of time.
-
27.
Calculate the Corrected TR-FRET Ratios on a per-well basis by subtracting the average TR-FRET ratio of the DMSO-containing wells at each time point.
-
28.
Plot the Corrected TR-FRET Ratios against time. Fit the data to a one phase decay model using Equation 11 below:
| (Equation 11) |
Where Y0 is the initial height of the curve, K is the rate constant (koff in this case) equal to the reciprocal of the x-axis units, and plateau is the plateau height of the curve (constrained to 0 since background has been subtracted).
TR-FRET assay to quantify endogenous BRD4 abundance in cell extract after degrader treatment
Timing: 2 days
This assay is used to quantify the abundance of endogenous BRD4 in cell extract after degrader treatment. The assay system consists of a primary rabbit anti-BRD4 antibody, a CoraFluor-1-labeled anti-rabbit Nano-Secondary, and JQ1-FITC.4 In the detection mix, JQ1-FITC is employed at a concentration much higher than its KD,app for endogenous BRD4 (100 nM used in assay, versus KD,app = 8.9 nM). This higher concentration of JQ1-FITC is used so that (i) maximum occupancy of BRD4 is achieved, resulting in a larger assay linear range, and (ii) effects from ligand competition from remaining PROTAC in the lysate are minimized. This protocol specifies the use of MDA-MB-231 cells; however, the protocol should be amenable to other adherent mammalian cell lines of interest. Furthermore, as the quantification assay concept is target-agnostic, if suitable primary antibodies and fluorescent tracers are identified for other targets of interest, a similar quantification strategy may be employed.
-
29.
Seed MDA-MB-231 cells at a density of 20,000 cells/well into a 96-well plate (Corning 3904 or similar) in 100 μL of cell culture medium.
-
30.
Allow the cells to adhere overnight at 37°C and 5% CO2.
-
31.
Dispense compounds (e.g., via HP D300) according to plate map in Figure 5. The standard dose-response range is from 2.5 μM to 0.26 nM (11-point dose-response, 2.5-fold dilution steps).
Note: While we have not observed meaningful edge effects in this assay, the plate map in Figure 5 can be adjusted to eliminate the outermost rows and columns of the plate if desired.
-
32.
Incubate cells with test compounds for 5 h at 37°C and 5% CO2.
Note: Different incubation times may be optimal for other target/PROTAC pairs.
-
33.
Gently aspirate medium from all wells using a multichannel pipette (e.g., ThermoFisher Matrix Equalizer 2034).
-
34.
Replace with fresh cell culture medium (150 μL/well) using a multichannel pipette.
-
35.
Incubate cells for 1 h at 37°C and 5% CO2 to wash out residual test compound.
-
36.
Gently aspirate medium from all wells using a multichannel pipette.
CRITICAL: If using cell culture medium containing phenol red, try to ensure that nearly all cell culture medium is aspirated during this step, as phenol red can interfere with downstream TR-FRET assay performance.
-
37.
Add 1× PBS (200 μL/well) using a multichannel pipette.
-
38.
Gently aspirate PBS from all wells using a multichannel pipette.
-
39.
Add freshly prepared, ice-cold Complete Lysis Buffer (60 μL/well) using a multichannel pipette.
-
40.
Shake plate at 1,000 rpm on an orbital shaker (e.g., Boekel Scientific Jitterbug, model 130000) at 20°C–22°C for 10 min.
-
41.Add freshly prepared 7× control detection mix and 7× complete detection mix (10 μL/well) using a multichannel pipette, according to the plate map in Figure 6.
-
a.7× control detection mix = 7 nM CoraFluor-1-labeled anti-rabbit Nano-Secondary, 700 nM JQ1-FITC, prepared in Dilution Buffer.
-
b.7× complete detection mix = 3.5 nM rabbit anti-BRD4 antibody, 7 nM CoraFluor-1-labeled anti-rabbit Nano-Secondary, 700 nM JQ1-FITC, prepared in Dilution Buffer.
-
a.
-
42.
Incubate the plate for 1 h at room temperature.
-
43.
Centrifuge the plate at 200 × g for 1 min.
-
44.
Transfer 30 μL of lysate (×2 TR-FRET replicates) into wells of a 384-well (e.g., Corning 3572) using either an adjustable electronic multichannel pipette (e.g., ThermoFisher Matrix Equalizer 2231; see Figure 7) or a standard handheld multichannel pipette (see Figure 8).
Note: The 96-well plates used for cell culture (Corning 3904) are clear-bottom plates, which are used so that cell health and density can be assess with a conventional bright-field microscope. These plates do not give optimal signal on a TR-FRET plate reader. Therefore, after cell lysis in the 96-well plate, the lysates are transferred to a white 384-well plate, which is optimal for TR-FRET readouts. This protocol may be adapted if cell culture of the desired cells is compatible with white, opaque 96- or 384-well plates, where the TR-FRET measurement can be performed directly in the same plate after cell lysis.
-
45.
Take the TR-FRET ratio on a per-well basis.
-
46.
Calculate the Normalized TR-FRET Ratios on a per-well basis using Equation 12 below:
| (Equation 12) |
-
47.Plot the Normalized TR-FRET Ratios against the log10 concentration of test compound.
-
a.In this assay, the Normalized TR-FRET Ratio is a direct measure of the abundance of BRD4 relative to DMSO-treated wells. Background signal (Normalized TR-FRET ratio of 0) is equal to 100% degradation.
-
a.
Figure 5.
Standard plate map for treatment of MDA-MB-231 cells with PROTACs in 96-well plates
Arrows indicate decreasing compound concentration for one compound in one biological replicate. In this format, 8 compounds can be fitted per 96-well plate.
Figure 6.
Standard plate map for addition of 7× control detection mix and 7× complete detection mix after cell lysis in 96-well plates
Figure 7.
Plate map for transferring lysate with detection mix from 96-well plate to 384-well plate using an adjustable electronic multichannel pipette
The pipette spacing on the adjustable multichannel pipettes can accommodate the difference in well spacing for 96- and 384-well plates.
Figure 8.
Plate map for transferring lysate with detection mix from 96-well plate to 384-well plate using standard handheld multichannel pipette
A standard handheld 8-tip multichannel pipette will fill every other well in a 384-well plate.
TR-FRET assay to quantify BRD4(BD1)-GFP and BRD4(BD2)-GFP abundance in cell extract after degrader treatment
Timing: 2 days
This assay is used to quantify the abundance of BRD4(BD1)-GFP and BRD4(BD2)-GFP in cell extract from HEK293T cells stably expressing either GFP-fusion protein after PROTAC treatment.8 The detection mix consists of a CoraFluor-1-labeled anti-GFP VHH.5 This protocol specifies the use of HEK293T cells stably expressing either BRD4(BD1)-GFP or BRD4(BD2)-GFP; however, the protocol should be amenable to other adherent mammalian cell lines of interest expressing other target proteins as GFP-fusions.
Note: For N-terminal GFP-fusion proteins, it should be ensured that there are no substantial background contributions from prematurely terminated translation products.
-
48.
Seed HEK293T cells stably expressing BRD4(BD1)-GFP or BRD4(BD2)-GFP at a density of 20,000 cells/well into a 96-well plate (Corning 3904 or similar) in 100 μL of cell culture medium.
-
49.
Allow the cells to adhere overnight at 37°C and 5% CO2.
-
50.
Dispense compounds (e.g., via HP D300) according to plate map in Figure 5. The standard dose-response range is from 2.5 μM to 0.26 nM (11-point dose-response, 2.5-fold dilution steps).
-
51.
Incubate cells with test compounds for 5 h at 37°C and 5% CO2.
-
52.
Gently aspirate medium from all wells using a multichannel pipette.
CRITICAL: If using cell culture medium containing phenol red, try to ensure that nearly all cell culture medium is aspirated during this step, as phenol red can interfere with downstream TR-FRET assay performance.
-
53.
Add 1× PBS (200 μL/well) using a multichannel pipette.
-
54.
Gently aspirate PBS from all wells using a multichannel pipette.
-
55.
Add freshly prepared, ice-cold Complete Lysis Buffer (60 μL/well) using a multichannel pipette.
-
56.
Shake plate at 1,000 rpm on an orbital shaker (e.g., Boekel Scientific Jitterbug, model 130000) 20°C–22°C for 10 min.
-
57.Add freshly prepared 7× control detection mix and 7× complete detection mix (10 μL/well) using a multichannel pipette, according to the plate map in Figure 6.
-
a.7× control detection mix = 28 nM CoraFluor-1-labeled anti-GST VHH, prepared in Dilution Buffer.
-
b.7× complete detection mix = 28 nM CoraFluor-1-labeled anti-GFP VHH, prepared in Dilution Buffer.
-
a.
-
58.
Incubate the plate for 1 h at room temperature.
-
59.
Centrifuge the plate at 200 × g for 1 min.
-
60.
Transfer 30 μL of lysate (×2 TR-FRET replicates) into wells of a 384-well (e.g., Corning 3572) using either an adjustable electronic multichannel pipette (see Figure 7) or a standard handheld multichannel pipette (see Figure 8).
-
61.
Take the TR-FRET ratio on a per-well basis.
-
62.
Calculate the Normalized TR-FRET Ratios on a per-well basis using Equation 12.
-
63.
Plot the Normalized TR-FRET Ratios against the log10 concentration of test compound.
-
64.
In this assay, the Normalized TR-FRET Ratio is a direct measure of the abundance of BRD4(BD1)-GFP or BRD4(BD2)-GFP relative to DMSO-treated wells. Background signal (Normalized TR-FRET ratio of 0) is equal to 100% degradation.
Expected outcomes
The TR-FRET assay procedures described here allow the user to readily determine critical parameters for the development of small-molecule degraders, including binary and ternary complex target engagement, binding kinetics, and cellular degradation of target proteins of interest. In the research paper associated with this protocol, we have used these assays to quantitatively profile over 60 distinct heterobifunctional degraders targeting different proteins of interest.
The raw data obtained from these TR-FRET experiments should be (i) a decrease in TR-FRET signal with increasing concentration of competing ligand in ligand displacement assays (Figure 1A), (ii) a decrease in TR-FRET signal with increasing concentration of competing ligand in KD,ternary assays (Figure 1B), (iii) a kinetic decrease in TR-FRET signal upon competing ligand addition in ternary complex off-rate experiments (Figures 1C and 1D), and (iv) a decrease in TR-FRET signal relative to DMSO-treated wells in quantification experiments measuring endogenous or GFP-tagged target proteins (Figures 1E and 1F).
Quantification and statistical analysis
Statistical details of experiments can be found in the main protocol text or figure legends. For cell-based assays, n biological replicates have been defined as independent cell treatments, performed at different times with biologically distinct samples. For biochemical assays, the n numbers denoted in the figure legends refer to the number of technical replicates used to calculate mean ± SD for a given data point within an experiment. No statistical methods were used to predetermine sample size and investigators were not blinded to outcome assessment. GraphPad Prism 10 was used for statistical analyses using default settings.
Limitations
While TR-FRET assays are well-suited for most average-sized proteins (e.g., 50–150 kDa), larger protein complexes may result in the TR-FRET donor and acceptor being placed too far apart for efficient FRET to occur, resulting in poor assay performance. Troubleshooting may be required to optimize TR-FRET assays for larger proteins, for instance by site-specific labeling informed by structural information (where available).
The cellular degradation assays described here were designed for use with adherent mammalian cell lines. For those cell lines that are not adherent, adaptation of these assays to 96-well plate-based formats may be difficult, as washing steps are required during the procedure. Non-adherent cell lines will require adaptation of the washing protocol. Nonetheless, for non-adherent cell lines, lysates produced from cell pellets of PROTAC-treated cells will still be amenable to the TR-FRET quantification strategies described here, so long as those pellets can be generated by the user in their own adapted protocol.
The cell-based TR-FRET assay protocol to measure endogenous proteins of interest is described here for BRD4. However, it will often be desirable to extend this methodology to other target proteins of interest. In our experience, the main limitation in applying the protocol to other proteins of interest is the availability of a suitable primary antibody directed against the target protein. In general, antibodies that are validated in immunoprecipitation or enzyme-linked immunosorbent assay (ELISA) are good starting points. Furthermore, in our experience, TR-FRET tracer ligands with affinities < 1 μM are generally able to be adapted to this quantification format. Optimization of the lysate-based quantification for other target proteins can be adapted from Payne et al.4
Troubleshooting
Problem 1
Low recovery of labeled protein conjugate after labeling with CoraFluor-1-Pfp or AF488-Tfp (“preparation of CoraFluor-1 and AF488-labeled antibodies and proteins” – steps 1–5).
Potential solution
Ensure that the sample is applied directly to the center of the MWCO Zeba spin desalting column resin bed. The inclusion of 0.05% TWEEN-20 in storage buffers also helps to maximize sample recovery in our experience.
Problem 2
No specific TR-FRET signal observed during saturation binding experiments (“determination of equilibrium dissociation constants (KD or KD,app) for fluorescent tracers by TR-FRET” – steps 6–10).
Potential solution
Ensure that correct instrument settings are used. For CoraFluor-1 and FITC/AF488 donor-acceptor systems, measure emission at 490 nm (Terbium) and 520 nm (FITC/AF488).
Check the performance of CoraFluor-1-labeled antibody/nanobody reagents. For instance, titrate His6-GFP against CoraFluor-1-labeled anti-His6 antibody, or AF488-labeled SjGST protein against CoraFluor-1-labeled anti-GST VHH or anti-GST antibody.
Optimize assay conditions including buffer, protein concentration, and incubation time.
Problem 3
No change in TR-FRET signal during KD,ternary experiments with His6-CRBN-DDB1 and target proteins (“determining ternary complex KD values (KD,ternary) and cooperativity factor alpha (α) values of test compounds with His6-CRBN-DDB1 and target proteins” – steps 10–19).
Potential solution
Ensure that the target protein does not have a His6-tag, as this will scavenge CoraFluor-1-labeled anti-His6 antibody from His6-CRBN-DDB1, disrupting the assay.
Problem 4
TR-FRET signal not above background in initial TR-FRET measurement during ternary complex off-rate experiments (“determining dissociation rate constant (koff) values for PROTAC-mediated ternary complexes between His6-CRBN-DDB1 and target proteins” – steps 20–28).
Potential solution
Optimize the concentration of His6-CRBN-DDB1 and the target protein.
In addition, optimize the concentration of PROTAC to be used. Usually, the optimal sensitivity will be at a concentration equal to the KD,ternary value of the PROTAC.
Problem 5
Cellular assays have low performance and/or high assay variance (“TR-FRET assay to quantify endogenous BRD4 abundance in cell extract after degrader treatment” and “TR-FRET assay to quantify BRD4(BD1)-GFP and BRD4(BD2)-GFP abundance in cell extract after degrader treatment” – steps 29–64).
Potential solution
Ensure that all media replacement and washing steps are performed slowly so that cells remain adherent during manipulation steps.
If using media containing phenol red, be sure to thoroughly remove the media from the cells during media removal and washing steps before TR-FRET measurements.
Resource availability
Lead contact
Further information and requests for resources and reagents generated in this study should be directed to the lead contact, Ralph Mazitschek (ralph@broadinstitute.org).
Technical contact
Questions about the technical specifics of performing the protocol should be directed to and will be answered by the technical contact, N. Connor Payne (npayne@hsph.harvard.edu).
Materials availability
All unique cell lines and reagents generated in this study are available from the lead contact upon request as long as stocks remain available.
Data and code availability
-
•
All data are available in the main text.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
His6-CRBN-DDB1 was a generous gift from Bristol Myers Squibb. Support from the Ono Pharma Foundation (C.M.W.), Blavatnik Biomedical Accelerator at Harvard University (C.M.W.), the National Institutes of Health (R01GM141406, C.M.W.; 1R01NS064983, R.M.), the National Science Foundation (1830941, R.M.), Japan Society for the Promotion of Science (S.I.), and National Science Foundation graduate research fellowship (N.C.P.) is gratefully acknowledged.
Author contributions
R.M. and C.M.W. supervised all aspects of the project. R.M., N.C.P., C.M.W., and S.I. conceived of the project. S.I. and N.C.P. synthesized compounds. N.C.P. and S.I. performed the experiments. N.C.P., S.I., C.M.W., and R.M. analyzed the data. N.C.P. drafted the manuscript, and all authors jointly discussed and edited the manuscript.
Declaration of interests
C.M.W. and S.I. are inventors of a patent application filed by Harvard University covering the chemical structures of cyclimids and their use. R.M. and N.C.P. are inventors on patent applications related to the CoraFluor TR-FRET technology used in this work.
Contributor Information
Christina M. Woo, Email: cwoo@chemistry.harvard.edu.
Ralph Mazitschek, Email: ralph@broadinstitute.org.
References
- 1.Ichikawa S., Payne N.C., Xu W., Chang C.F., Vallavoju N., Frome S., Flaxman H.A., Mazitschek R., Woo C.M. The cyclimids: Degron-inspired cereblon binders for targeted protein degradation. Cell Chem. Biol. 2024 doi: 10.1016/j.chembiol.2024.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne N.C., Kalyakina A.S., Singh K., Tye M.A., Mazitschek R. Bright and stable luminescent probes for target engagement profiling in live cells. Nat. Chem. Biol. 2021;17:1168–1177. doi: 10.1038/s41589-021-00877-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng Y., Prusoff W.H. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 4.Payne N.C., Maksoud S., Tannous B.A., Mazitschek R. A direct high-throughput protein quantification strategy facilitates discovery and characterization of a celastrol-derived BRD4 degrader. Cell Chem. Biol. 2022;29:1333–1340.e5. doi: 10.1016/j.chembiol.2022.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Payne N.C., Mazitschek R. Tiny Titans: Nanobodies as Powerful Tools for TR-FRET Assay Development. Analysis Sensing. 2022;2 [Google Scholar]
- 6.Lin W., Li Y., Min J., Liu J., Yang L., Lee R.E., Chen T. Development of BODIPY FL Thalidomide As a High-Affinity Fluorescent Probe for Cereblon in a Time-Resolved Fluorescence Resonance Energy Transfer Assay. Bioconjug. Chem. 2020;31:2564–2575. doi: 10.1021/acs.bioconjchem.0c00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller G.W., Chen R., Huang S.Y., Corral L.G., Wong L.M., Patterson R.T., Chen Y., Kaplan G., Stirling D.I. Amino-substituted thalidomide analogs: potent inhibitors of TNF-alpha production. Bioorg. Med. Chem. Lett. 1999;9:1625–1630. doi: 10.1016/s0960-894x(99)00250-4. [DOI] [PubMed] [Google Scholar]
- 8.Nowak R.P., DeAngelo S.L., Buckley D., He Z., Donovan K.A., An J., Safaee N., Jedrychowski M.P., Ponthier C.M., Ishoey M., et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat. Chem. Biol. 2018;14:706–714. doi: 10.1038/s41589-018-0055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jarmoskaite I., AlSadhan I., Vaidyanathan P.P., Herschlag D. How to measure and evaluate binding affinities. Elife. 2020;9 doi: 10.7554/eLife.57264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu J., Qian Y., Altieri M., Dong H., Wang J., Raina K., Hines J., Winkler J.D., Crew A.P., Coleman K., Crews C.M. Hijacking the E3 Ubiquitin Ligase Cereblon to Efficiently Target BRD4. Chem. Biol. 2015;22:755–763. doi: 10.1016/j.chembiol.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winter G.E., Buckley D.L., Paulk J., Roberts J.M., Souza A., Dhe-Paganon S., Bradner J.E. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348:1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry D.W., Harvey P.J., Keller P.R., Elliott W.L., Meade M., Trachet E., Albassam M., Zheng X., Leopold W.R., Pryer N.K., Toogood P.L. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol. Cancer Ther. 2004;3:1427–1438. [PubMed] [Google Scholar]
- 13.Douglass E.F., Jr., Miller C.J., Sparer G., Shapiro H., Spiegel D.A. A comprehensive mathematical model for three-body binding equilibria. J. Am. Chem. Soc. 2013;135:6092–6099. doi: 10.1021/ja311795d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gadd M.S., Testa A., Lucas X., Chan K.H., Chen W., Lamont D.J., Zengerle M., Ciulli A. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farnaby W., Koegl M., Roy M.J., Whitworth C., Diers E., Trainor N., Zollman D., Steurer S., Karolyi-Oezguer J., Riedmueller C., et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 2019;15:672–680. doi: 10.1038/s41589-019-0294-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roy M.J., Winkler S., Hughes S.J., Whitworth C., Galant M., Farnaby W., Rumpel K., Ciulli A. SPR-Measured Dissociation Kinetics of PROTAC Ternary Complexes Influence Target Degradation Rate. ACS Chem. Biol. 2019;14:361–368. doi: 10.1021/acschembio.9b00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imaide S., Riching K.M., Makukhin N., Vetma V., Whitworth C., Hughes S.J., Trainor N., Mahan S.D., Murphy N., Cowan A.D., et al. Trivalent PROTACs enhance protein degradation via combined avidity and cooperativity. Nat. Chem. Biol. 2021;17:1157–1167. doi: 10.1038/s41589-021-00878-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
-
•
All data are available in the main text.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.