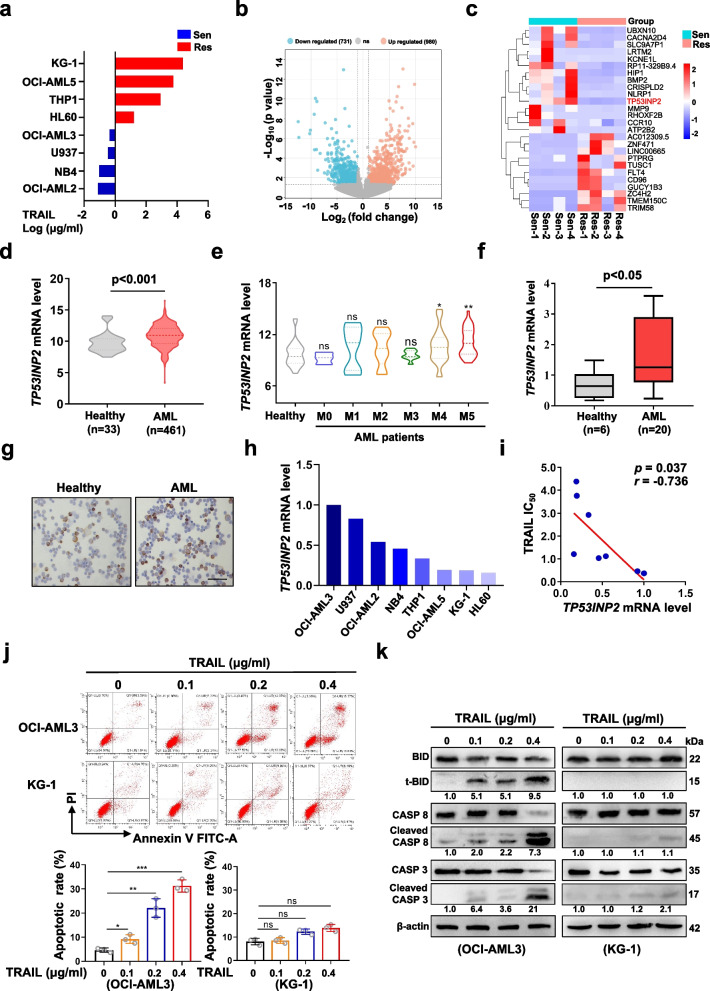
Fig. 1.
The association between TP53INP2 and the sensitivity of AML cells to TRAIL treatment was identified by high-throughput library screening. a The IC50 was estimated by CCK-8 with the concentration of TRAIL that resulted in 50% cell viability for 8 AML cells. The AML cells were divided into TRAIL-Sen and TRAIL-Res cells according to the IC50 against TRAIL. b Volcanic plot of the DEGs between TRAIL-Sen and TRAIL-Res AML cells. c Heatmap of the top 20 DEGs between TRAIL-Sen and TRAIL-Res AML cells. d Comparison of TP53INP2 mRNA levels in AML patients and healthy donors in Beat AML databases. e Comparison of TP53INP2 mRNA levels in various subtypes of AML patients in Beat AML databases. f-g qRT-PCR (f) and IHC staining (g) analyses of TP53INP2 levels in primary AML blasts and healthy donors (Scale bar: 50 μm). The upper, center, and lower line of the boxplot indicates 75%, 50%, and 25% quantile, respectively. Healthy donors served as controls in (d-g). h The expression of TP53INP2 in AML cells from the Cancer Cell Line Encyclopedia database. The ratio of TP53INP2/β-actin in OCI-AML3 cells was set to 1.0. i Pearson’s correlation analysis of TP53INP2 expression and the IC50 against TRAIL in AML cells. j FCM analysis (j, upper) and quantification (j, lower) of apoptotic cells in OCI-AML3 and KG-1 cells treated with 0–0.4 μg/ml TRAIL for 48 h. The cells incubated in drug-free medium served as controls. k Western blot analysis of the indicated apoptosis-related proteins. β-actin was used as a loading control. The quantification of protein levels was shown below the protein bands. The data are representative of at least three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, ns, not significant