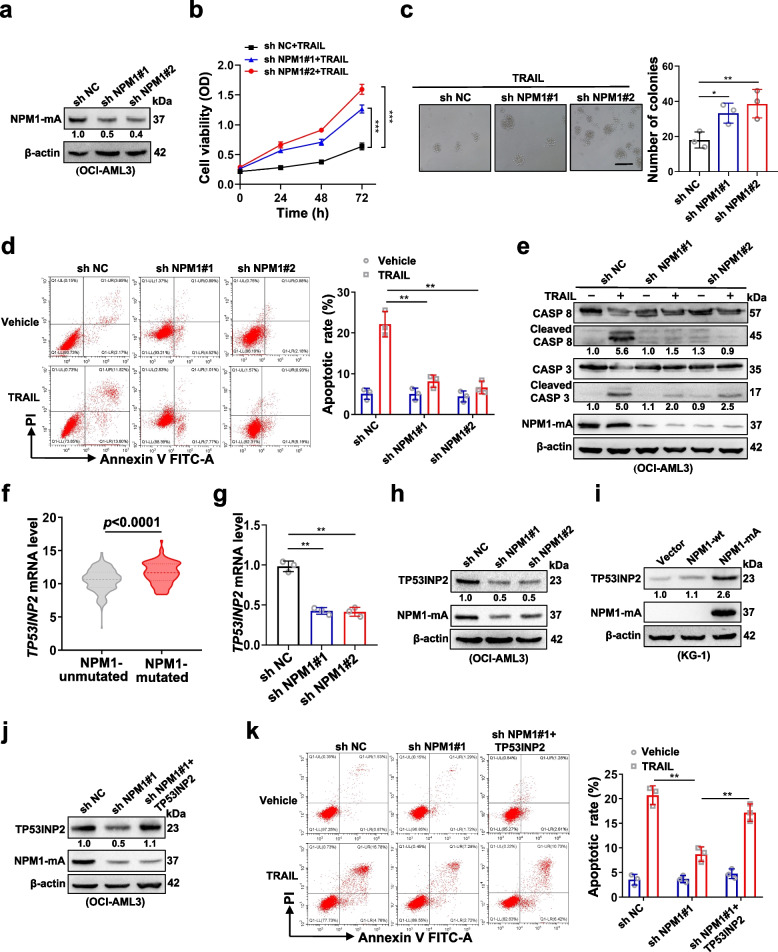
Fig. 4.
The effect of TP53INP2 on TRAIL sensitivity may be regulated by the mutant NPM1. a Western blot analysis of NPM1-mA in OCI-AML3 cells infected with two sh RNAs targeting NPM1. b CCK-8 analysis (n = 3) of cell viability in the cells treated with 100 ng/ml TRAIL for 0–72 h. c Colony formation assay was performed in OCI-AML3 cells. The representative images and quantitative data from three independent experiments were shown in (c, left) and (c, right), respectively. (Scale bar: 50 μm). d FCM analysis (d, left) and quantification (d, right) of apoptotic cells in the cells. e Western blot analysis of the indicated apoptosis-related proteins. f TP53INP2 levels in NPM1-mutated AML patients, compared with NPM1-unmutated AML patients, were identified from Beat AML databases. g qRT-PCR analysis of TP53INP2 mRNA levels in NPM1-mA-silenced OCI-AML3. h Western blot analysis of TP53INP2 and NPM1-mA in OCI-AML3 cells with two sh NPM1. i Western blot analysis of TP53INP2 and NPM1-mA in KG-1 cells with the NPM1-wt and NPM1-mA plasmids. j Western blot analysis of TP53INP2 and NPM1-mA. In (a, e, h-g), β-actin was used as a loading control, and the quantification of protein levels was shown below the protein bands. k FCM analysis (k, left) and quantification (k, right) of apoptotic cells in OCI-AML3 cells with sh NPM1 before transfection with the HA-TP53INP2 plasmids. The cells transfected with shNC or vectors served as controls. The data are representative of at least three independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001