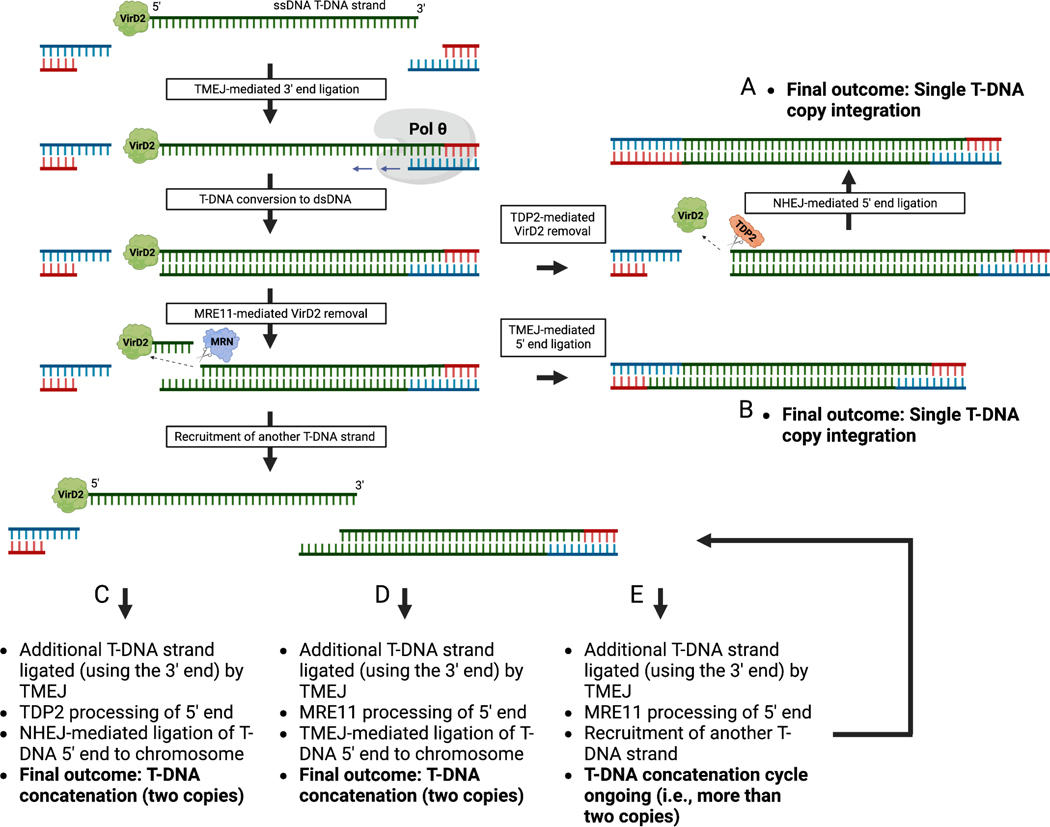
Extended Data Figure 7. Model of T-DNA concatenation.
The model builds on the T-DNA integration model from Kralemann et al., 20226. Chromosomal capture of a T-DNA strand 3’ end is mediated by TMEJ. After conversion to a double-stranded T-DNA intermediate, capture of the T-DNA 5’ end is accomplished by removal of the Agrobacterium protein VirD2 by TDP2 or MRE11. TDP2-mediated removal of VirD2 creates blunt-ended DNA at the T-DNA 5’ end that is ligated to the chromosome by NHEJ. In contrast, MRE11, acting as part of the MRN complex (MRE11-RAD50-NBS1; loaded on DNA by RAD1725), removes VirD2 by cutting the T-DNA internally, generating a staggered end at the T-DNA 5’ end. TDP2/NHEJ activity leads to a single T-DNA copy integration (outcome A), while MRE11/TMEJ activity leads to multiple outcomes (B-E), with the simplest one being chromosomal capture of the T-DNA 5’ end (outcome B). Alternatively, the staggered T-DNA 5’ end can facilitate recruitment of additional T-DNA strands for ligation, leading to concatenation. Capture of the 5’ end of an additional T-DNA strand by TDP2/NHEJ instead of MRE11/TMEJ is more likely to terminate the concatenation cycle. In this model, T-DNA features like DNA repeats may increase concatenation levels by increasing the number of available T-DNA strands for integration, and/or their accessibility. ssDNA: single-stranded DNA. dsDNA: double-stranded DNA. Created with BioRender.com.