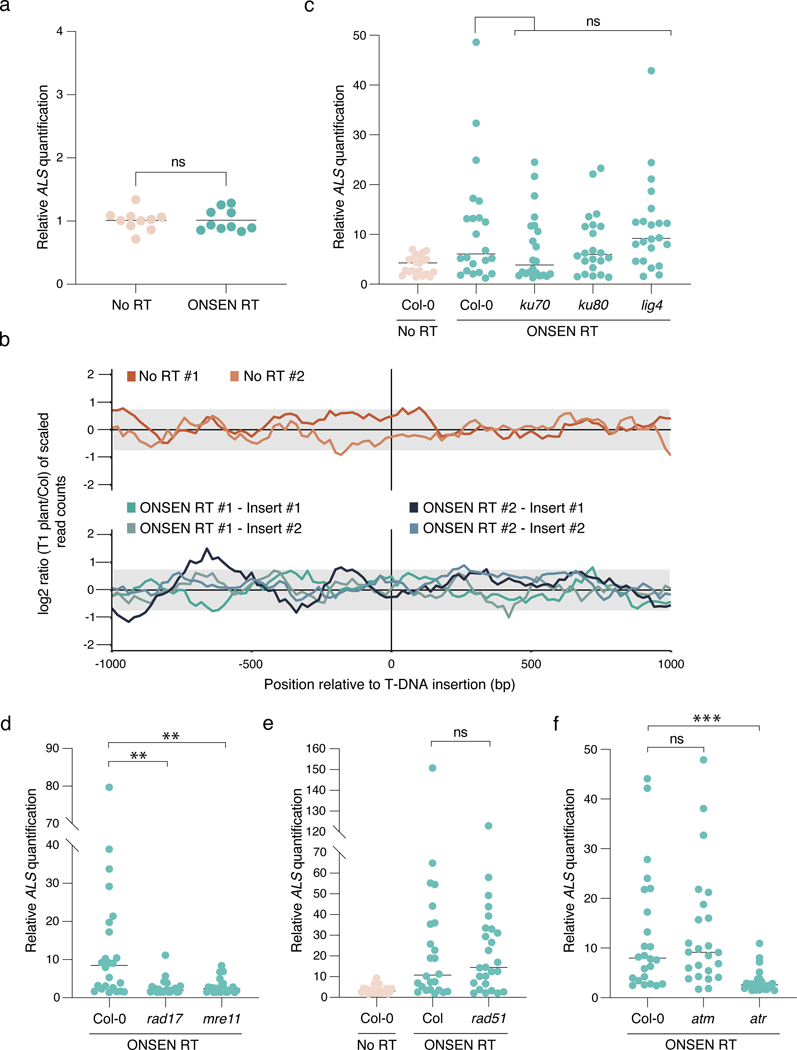
Figure 3. T-DNA concatenation is dependent on DNA repair.
a. DNA-qPCR of ALS in No RT- and ONSEN RT-transformed Agrobacterium. Each dot represents a liquid culture initiated from an independent colony (n = 20). Horizontal bars indicate the mean. ns = not significantly different (two-tailed Mann-Whitney U test). b. log2 ratio between the number of reads corresponding to the genomic regions surrounding the T-DNA insertion sites compared to Col-0 in 20 bp bins. The grey box represents the interval where 95% of copy number values from the Arabidopsis genome are present. c-f. DNA-qPCR of ALS in mutant backgrounds of the (c) NHEJ, (d) TMEJ, and (e) HR pathways, and of (f) DNA damage-induced kinases. Each dot represents an individual T1 plant (n = 24 for rad17, mre11, atm and atr, n = 22 for ku70, ku80 and lig4 and n = 26 for rad51). Horizontal bars indicate the median. Prad17 = 0.00233, Pmer11 = 0.00438 (Kruskal-Wallis ANOVA followed by Dunn’s test), Prad51 = 0.00233 (two-tailed Mann-Whitney U test), Patr = 0.00011 (Kruskal-Wallis ANOVA followed by Dunn’s test), ns = not significantly different. ns, P > 0.05; **, P < 0.01; ***, P < 0.001.