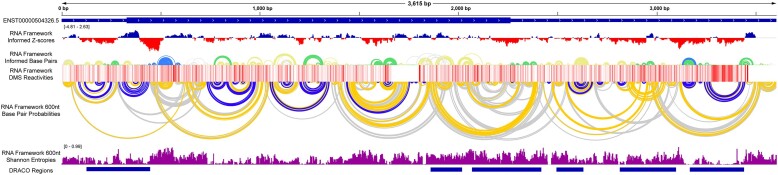
Figure 3.
All displayed data tracks were generated from RNA Framework informed predictions of AR-V7 and are presented as seen in the Integrative Genomics Viewer (IGV). The AR-V7 transcript cartoon is displayed at the top for spatial orientation of all other data. Below the transcript cartoon is the RNA Framework informed ScanFold z-scores with positive values in blue and negative values in red. Below the z-scores, the first arc diagram represents the RNA Framework informed ScanFold predicted base pairs with z-score >0, >–1, >–2 and ≤2 color in white/gray, yellow, green and blue respectively. Below the ScanFold arc diagram, the DMS reactivities are shown as a heat map on a scale of 0–1 where 0 is white, 1 is dark red, and intermediate values are shades of red. Below the DMS reactivity heat map, the second arc diagram represents the base pair probabilities calculated from an minimum free energy fold using RNA Framework reactivities and a 600 nt max base pair span constraint, with probabilities >80%, 30–80% and 10–30% displayed in blue, gold and gray respectively. Comparison of the ScanFold and base pair probability arc diagrams demonstrates the general agreement between low z-score structures and highly probable base pairings. Below the base pair probability arc diagram, the purple bar graph represents the Shannon entropies calculated using RNA Framework reactivities and a 600 nt max base pair span constraint. The blue bars at the bottom represent dynamic regions identified by DRACO.