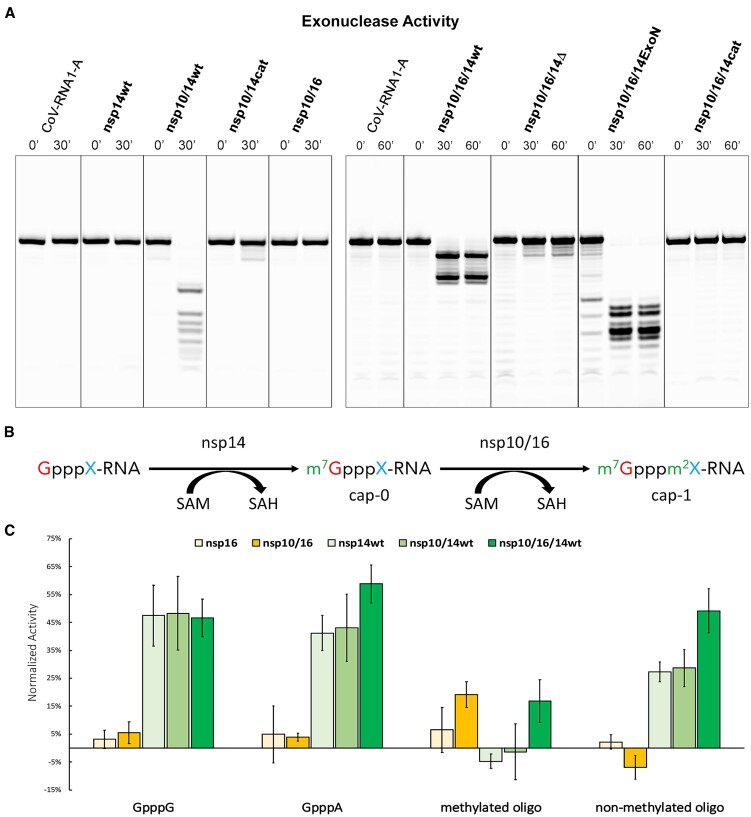
Figure 5.
The formation of nsp10/16/14wt heterotrimer modulates the ribonuclease, but not methyltransferase activities. (A) Exonuclease activity. CoV-RNA1-A degradation profiles of nsp14wt, nsp10/14 (wt, cat), nsp10/16, with 0- and 30-min incubation, and profiles of nsp10/16/14 (wt, Δ, ExoN, and cat) from 0-, 30- and 60-min incubation. Merged from Supplementary Figure S5A–C. (B) Schematic view of mRNA methylation. X represents a nascent nucleotide (adenine (A) or guanine (G)). m7 represents methylation of the first guanine at position N7 by nsp14. m2 indicates 2′-O methylation of the nascent mRNA nucleotide. (C) Methyltransferase activity of tested complexes. The results were normalized at SAH concentration. All experiments were performed in duplicate. Average values with error bars (SD) are shown. The Y-axis was normalized to the negative control in which the volume of the inhibitor was replaced with buffer supplemented with DMSO.