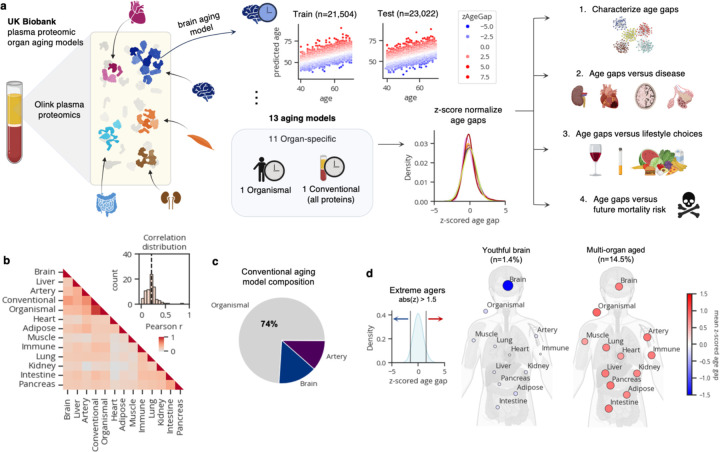
Figure 1. Plasma protein-derived organ age estimates in the UK Biobank.
a, Study design to estimate organ-specific biological age from plasma proteomics data in the UK Biobank. A protein was called organ-specific if the gene encoding the protein was expressed at least four-fold higher in one organ compared to any other organ in the GTEx organ bulk RNA-seq atlas. Organ-specific protein sets were used to train LASSO chronological age predictors. Samples from 10/21 centers (n=21,504) were used for training and the remaining samples (n=23,022) were used for testing. An ‘organismal’ model, which was trained on the levels of non-organ-specific (organ-shared) proteins, and a ‘conventional’ model, which was trained on all proteins on the Olink assay, were also developed and assessed. Model age gaps were calculated and then z-score normalized per organ to allow for direct comparisons across organs. Age gaps were characterized (Fig. 1), and tested for associations with disease risk (Fig. 2), modifiable lifestyle choices (Fig. 3), and mortality risk (Fig. 4).
b, Pairwise correlation of organ age gaps from all samples. Inset histogram shows the distribution of all pairwise correlations, with the dotted line representing the mean.
c, A Lasso regression model was used to predict conventional age based on organ ages and organismal age. Organismal, brain, and artery ages were sufficient to predict conventional age with r2=0.97. Relative weights are shown as a pie chart.
d, Extreme agers were defined by a 1.5 standard deviation increase or decrease in at least one age gap. The mean organ age gaps of extremely youthful brain agers and accelerated multi-organ agers are shown.