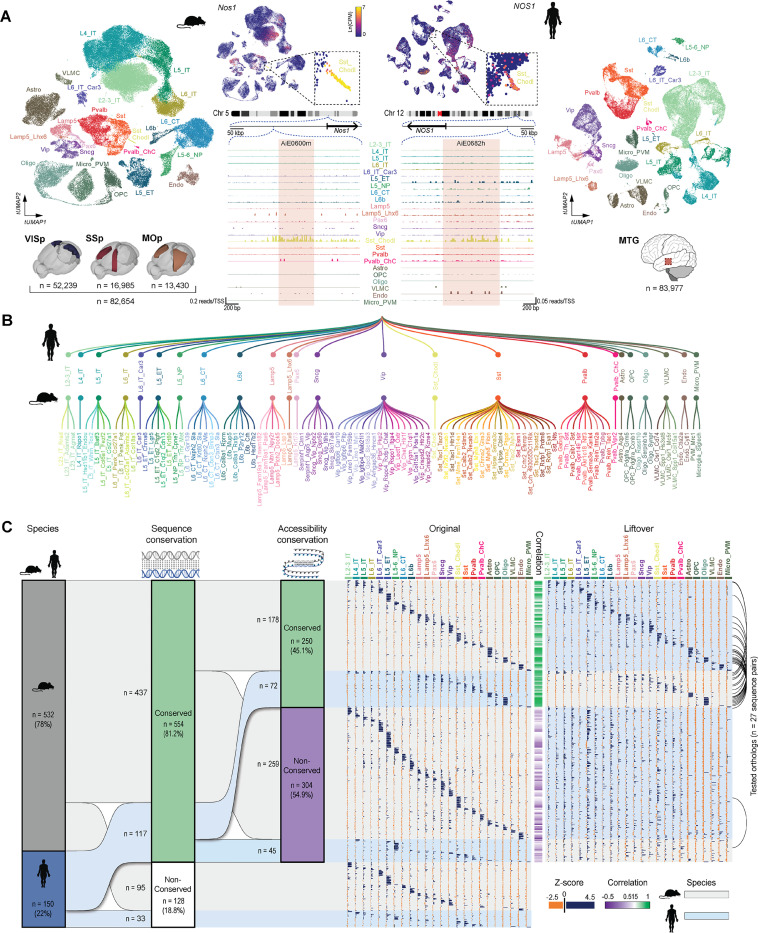
Figure 2: Selection of putative enhancer sequences.
A. Mouse and human single-nucleus transcriptomes obtained from single-nucleus multiomes represented in a transcriptomic UMAP and labeled according to cortical cell subclasses. Mouse nuclei were collected from the primary visual (VISp), somatosensory (SSp) and motor (MOp) cortices; human nuclei were collected from the middle temporal gyrus (MTG). Numbers of nuclei included in the analysis are represented by ‘n’. B. Simplified representation of the unified mouse-human taxonomy of cortical cell subclasses, along with the cluster-level taxonomy for mouse only. C. Summary of all ‘native’ putative enhancer sequences tested (n = 682), divided by the genome of origin, followed by cross-species conservation of sequence and accessibility (left). The modified sequences produced by concatenation are not included. The ‘Original’ and ‘Liftover’ plots show relative accessibility of all individual sequences in each subclass in their respective species, alongside the relative accessibility of its orthologous liftover sequence in the other species, respectively. Correlation between the original and liftover accessibility data is shown as a green/purple heatmap in the middle of these plots. Black arcs on the very right indicate instances where orthologs from both species were tested.