Figure 3: A pipeline for enhancer AAV screening in the mouse brain.
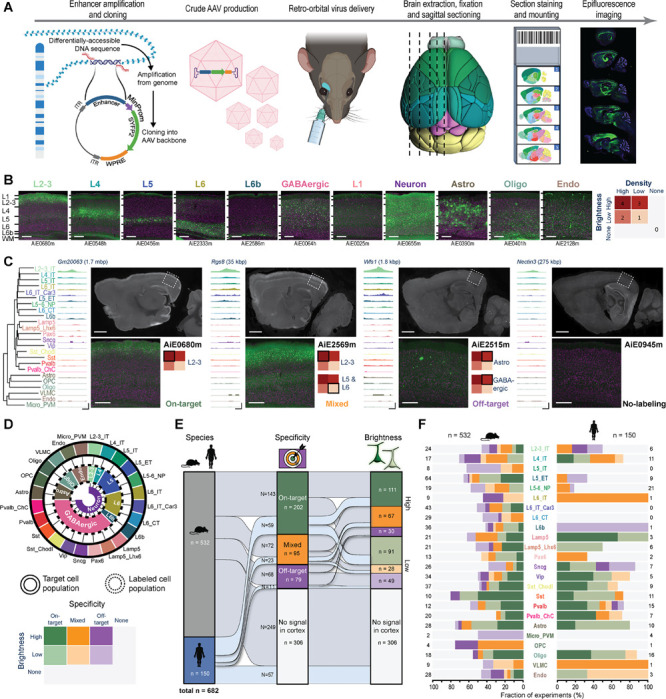
A. Graphical summary of the enhancer screening pipeline. B. Representative images of the visual cortex, showing examples of 11 categories of visually distinguishable cortical populations used to evaluate the labeling pattern of each enhancer AAV. Individual enhancer IDs are shown beneath each image (left), and a scoring matrix for evaluating brightness of the fluorescent signal and the labeling density, compared to the density expected for each category (right). C. Representative tracks of chromatin accessibility for four individual enhancers targeting across the cortical subclasses, demonstrating differential accessibility in L2–3_IT, alongside representative epifluorescence image sets, showing the resulting labeling pattern, and the score given to each in VISp. The closest L2–3_IT marker gene is shown above each set of tracks, along with the distance from the enhancer to its TSS. Scale bars below the tracks represent 100 bp (horizontal) and 0.3 RPKM/cell (vertical). D. Schematic describing the approach used to determine target specificity, according to the alignment between the TCP and LCP (top left) and a matrix used for classifying all enhancers, based on a combination of their target specificity and signal brightness (bottom left). E. Summary plot showing performance of all tested enhancers (n = 682) according to the categories (right). > 50% of enhancers (n = 376) exhibited signal in the cortex, ~43% were putatively on-target or had mixed (on- and off-target) labeling, and ~30% were putatively on-target (202/682). F. Proportion of enhancers in each of the categories specified in (D), according to their genome of origin (top, n = 150 for human and 532 for mouse) and TCP. Numerical values represent the number of tested enhancers in each column. For images, scale bars for full section and expanded views are 1.0 and 0.2 mm, respectively.
