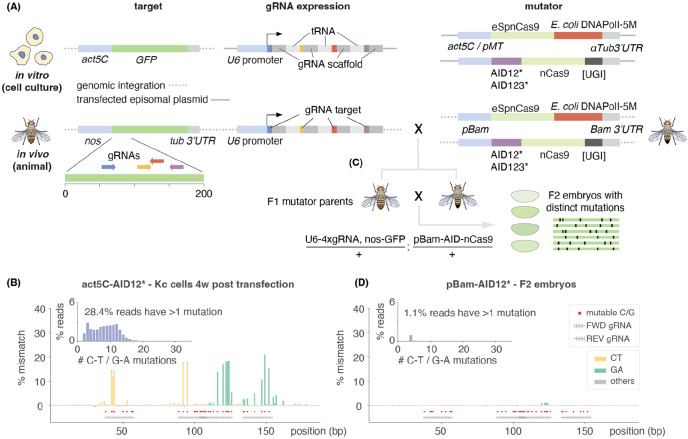
Figure 2. Human AID derived variants introduce C→T mutations at high rates at gRNA target sites in cell culture, but not in animals.
(A) Plasmids used in experiments in cell culture and flies to mutagenize GFP in the genome of Kc cells or fly embryos using 4 gRNAs and nCas9 fused to mutagenic proteins AID or EvolvR. Different AID variants, promoters and 3’UTRs were tested. gRNAs were expressed off of U6 ubiquitous promoters. (B) Barplots showing mismatches to GFP classified by base substitution generated by AID12* in Kc-nls-GFP cells 4 weeks post-transfection. Most mismatches are C→T (yellow) on forward gRNAs or G→A (green) on reverse gRNAs. See Fig. S2 for other conditions (C) In embryos, the mutagenic enzymes were expressed in the germline from a pBam promoter and crossed to flies containing the same U6-4xgRNA construct used in cell culture as well as GFP. F2 embryos were collected, and GFP amplified to then be sequenced using Nanopore sequencing. (E) Barplots showing mismatches to GFP classified by base substitution generated by AID12* in F2 embryos following one generation of germline mutations. Few G→A (green) mutations were detected. In (C) and (E) red dots mark all “mutable bases” (Cs on FWD gRNAs and Gs on REV gRNAs). Insets in each plot show the distribution of the number of C→T or G→A mismatches per read on these mutable bases, after subtracting the number of mismatches observed in control samples (see methods).