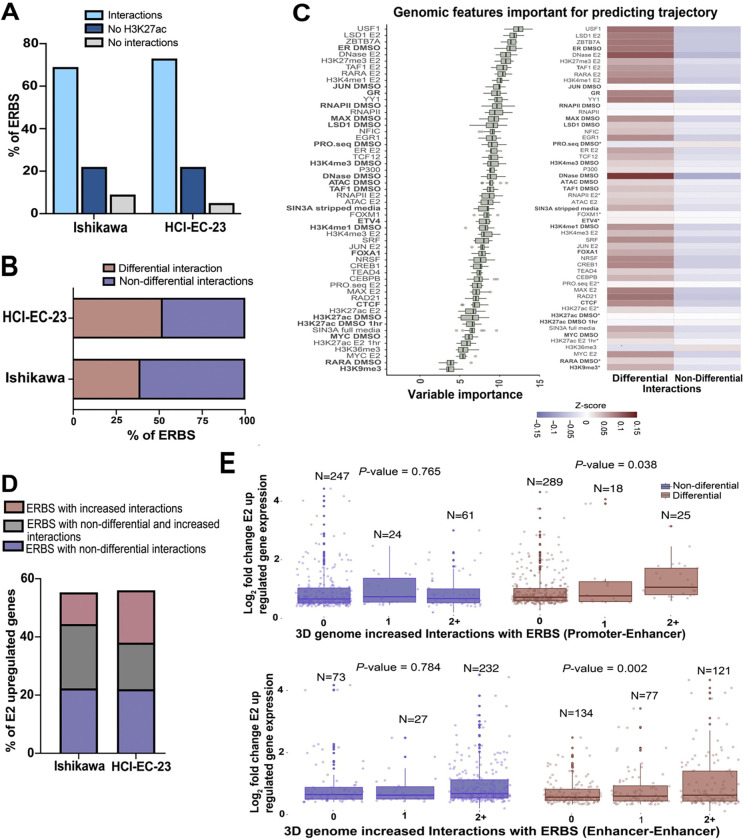
Figure 3. ERBS exhibit distinctive features when involved in differential 3D genome interactions.
(A) The bar graph portrays all ERBS associated with 3D genome interactions and their classifications in endometrial cell lines. (B) Of the ERBS found in 3D genome interactions (light blue in panel A), the bar graph indicates the percentage that anchors differential 3D genome interactions. (C, left) A ranking of genomic features based on their importance for ERBS associated with differential or non-differential 3D genome interactions (exhibited in panel B) is shown. Datasets shown in bold were performed in the absence of ER activation. (C, right) Heatmap displays the average signal intensity for top-ranked genomic features clustered by ERBS with differential or non-differential 3D genome interactions. Each feature’s distributions were significantly different between groups based on a Kruskal Wallis test (False Discovery Rate < 0.05) with the exception of features marked with an *. (D) Bar graph shows the percentage of E2 upregulated genes associated with ERBS in differential or non-differential 3D genome interactions (from panel B) within 100 kb of their TSS. (E) Box plots show fold change for E2 upregulated genes with at least one ERBS within 100 kb of their TSS in Ishikawa cells split by the number of non-differential (left, blue) or differential (right, red) Promoter–enhancer (top) or Enhancer–enhancer (bottom) chromatin loops anchored by ERBS. P-values are calculated as t-tests of the linear regression coefficients being different from 0.