Figure 2. Free energy landscape of PKA-C obtained from replica-averaged metadynamics (RAM) simulations.
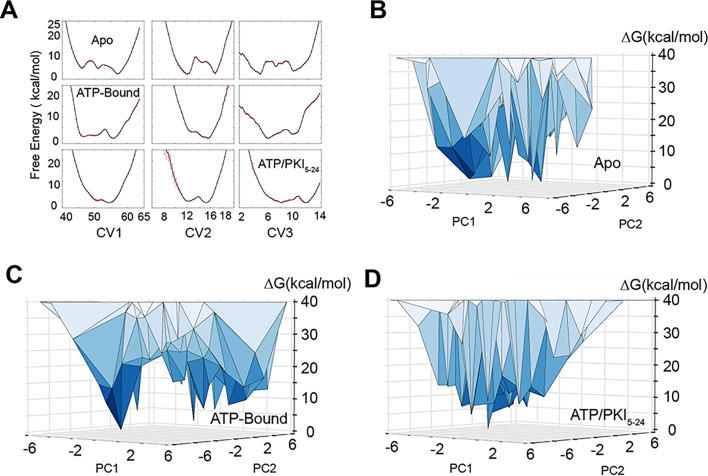
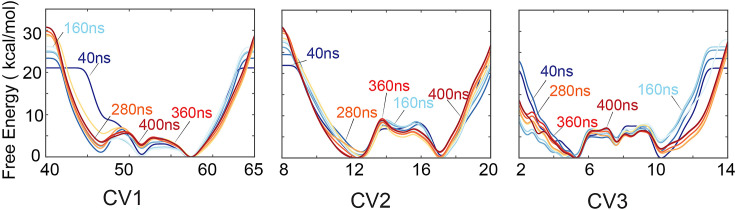
(A) Convergence of the bias deposition along the first three collective variables (CVs). The free energy (expressed in kcal/mol) of the different CVs was averaged over the last 100 ns of RAM simulations. The standard deviations are reported as red error bars. (B–D) Free energy landscape along the first two principal components (PC1 and PC2) of PKA-C in the apo, ATP-, and ATP/PKI-bound forms. PC1 and PC2 are projected from the first three CVs. The vertices represent conformational states. In the apo form, multiple states have comparable free energy with ΔG < 5 kcal/mol, whereas in the binary form, fewer states have ΔG < 5 kcal/mol. For the ternary form only a major ground state is populated.
Figure 2—figure supplement 1. Illustration of the collective variables (CVs) used in the replica-averaged metadynamics (RAM) simulations.
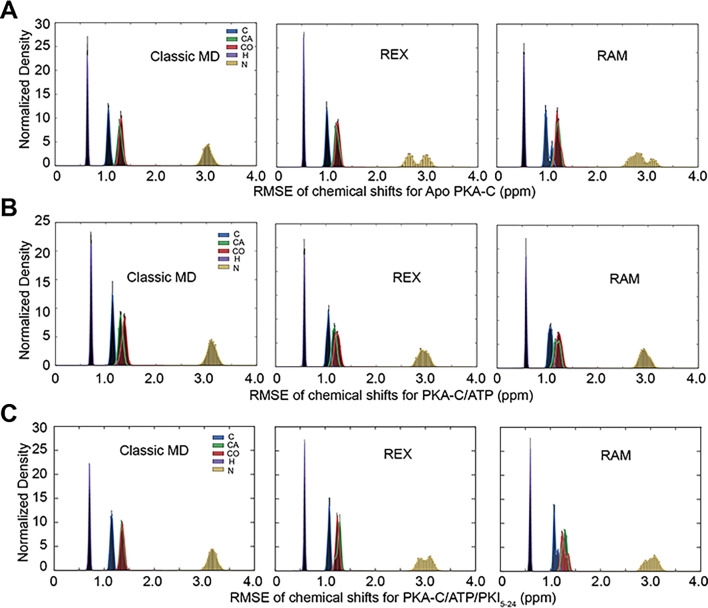
Figure 2—figure supplement 2. Distribution of the root-mean-square-error (RMSE) of the chemical shifts (CSs) for the different simulation schemes.
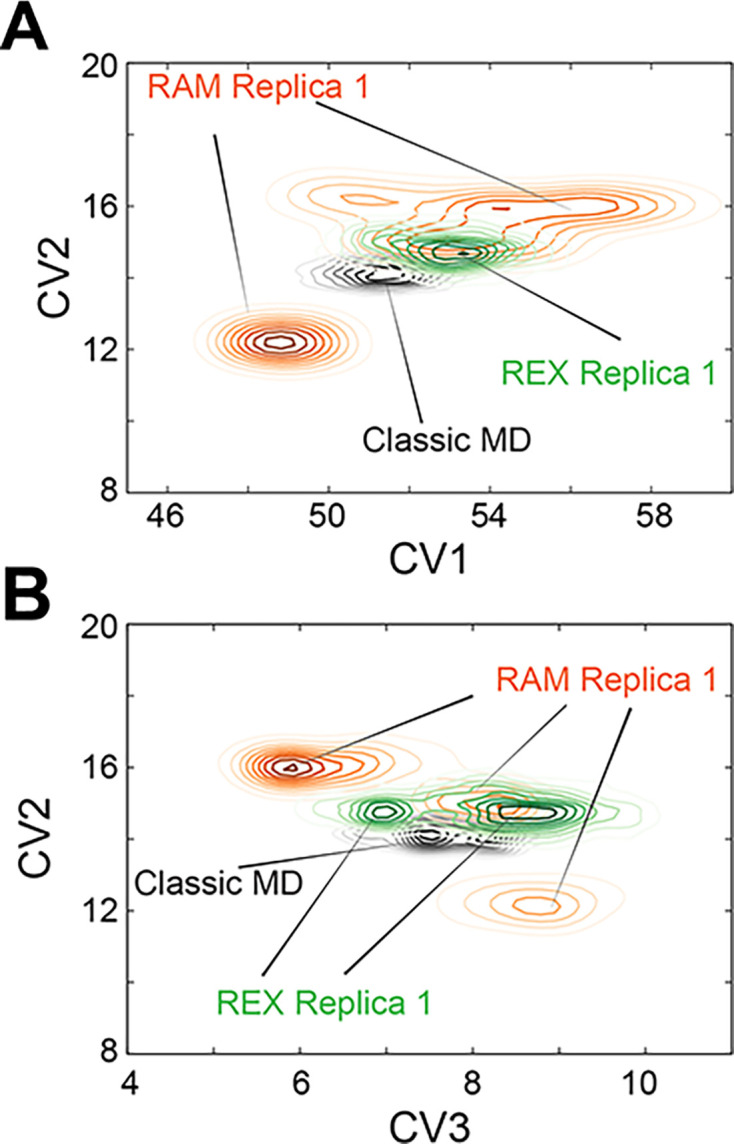
Figure 2—figure supplement 3. Replica-averaged metadynamics (RAM) simulations explore a larger conformational space than standard MD and replica exchange (REX) simulations.