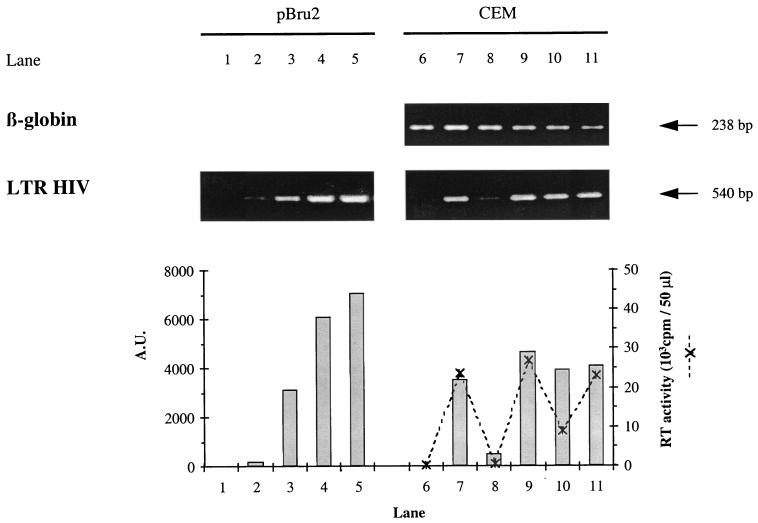
FIG. 5.
Estimation of HIV proviral DNA in CEM cells infected with the HIV-1 LAI CEM-adapted virus. At 24 h after infection, DNA was extracted from uninfected CEM (lane 6), infected CEM alone (lane 7), infected CEM cultivated with 5 μM AZT (lane 8), with supernatant from a 9-day-old culture of PHA-activated CD4+ cells from the EBV-positive donor (lane 9), with supernatant from a 9-day-old culture of antigen-stimulated 1.6-EBV line (lane 10), and with 1.6-EBV line cloning mix (lane 11). The quality of DNA extracts was monitored with a β-globin-specific PCR. A fourfold range of dilutions of the pBru2 plasmid, encoding the full-length HIV-1 Bru genome, from 1 × 105 to 6.4 × 106 copies per sample (lanes 2 to 5) was used as a calibration curve. Lane 1 corresponds to the negative control containing no DNA. The products obtained after the first template switching of the reverse transcription were amplified by a PCR-specific for the complete HIV-1 LTR. PCR products were quantified by using NIH 1.61 software, and their intensities were expressed as arbitrary units (A.U.) on a histogram. For CEM cells (lanes 6 to 11), the RT activity in supernatant fluids was expressed as the cpm/50 μl at the peak of viral replication (day 4, ---✻---).