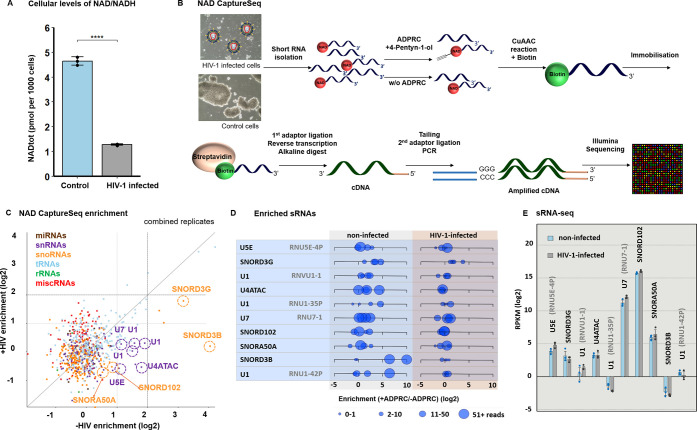
Figure 1.
Changes in NAD RNA capping upon HIV-1 infection. (A) Levels of free NAD/NADH in control and HIV-1-infected cells (measured in biological triplicates and technical duplicates, t-test). (B) Scheme of the NAD captureSeq protocol applied to control and HIV-1-infected cells. (C) Scatter plot showing the NAD captureSeq enrichment (+ADPRC vs −ADPRC) in control (X-axis) and HIV-1-infected samples (Y-axis), showing average values across replicates. Highlighted sRNAs passed the criteria listed in Figure S1B, i.e., change in NAD cap abundance upon HIV-1 infection, having specific enrichment in +ADPRC samples, sufficient read count and consistency across replicates; these sRNAs are visualized in more detail for individual replicates in panel (D). (D) Enrichment of RNAs in NAD captureSeq analysis enriched in control cells versus HIV-1-infected cells, matching highlighted sRNAs in Figure 1C. Blue circles show individual replicates; the size of the circle corresponds to the number of mapped reads. (E) sRNA–seq analysis of identified candidate sRNAs in control and HIV-1-infected cells (prepared in biological triplicates).