Abstract
Human herpesvirus 6 (HHV-6) and HHV-7 are closely related betaherpesviruses that encode a number of genes with no known counterparts in other herpesviruses. The product of one such gene is the HHV-6 glycoprotein gp82-105, which is a major virion component and a target for neutralizing antibodies. A 1.7-kb cDNA clone from HHV-7 was identified which contains a large open reading frame capable of encoding a predicted primary translational product of 468 amino acids (54 kDa) with 13 cysteine residues and 9 potential N-linked glycosylation sites. This putative protein, which we have termed gp65, was homologous to HHV-6 gp105 (30% identity) and contained a single potential membrane-spanning domain located near its amino terminus. Comparison of the cDNA sequence with that of the viral genome revealed that the gene encoding gp65 contains eight exons, spanning almost 6 kb of the viral genome at the right (3′) end of the HHV-7 genome. Northern (RNA) blot analysis with poly(A)+ RNA from HHV-7-infected cells revealed that the cDNA insert hybridized to a single major RNA species of 1.7 kb. Antiserum raised against a purified, recombinant form of gp65 recognized a protein of roughly 65 kDa in sucrose density gradient-purified HHV-7 preparations; treatment with PNGase F reduced this glycoprotein to a putative precursor of approximately 50 kDa. Gp65-specific antiserum also neutralized the infectivity of HHV-7, while matched preimmune serum did not do so. Finally, analysis of the biochemical properties of recombinant gp65 revealed a specific interaction with heparin and heparan sulfate proteoglycans and not with closely related molecules such as N-acetylheparin and de-N-sulfated heparin. At least two domains of the protein were found to contribute to heparin binding. Taken together, these findings suggest that HHV-7 gp65 may contribute to viral attachment to cell surface proteoglycans.
Human herpesvirus 6 (HHV-6) is a ubiquitous T-lymphotropic betaherpesvirus that was first discovered in 1986 (38) which has been etiologically linked to acute febrile illnesses in young children, including exanthem subitum (35, 53). A closely related virus, designated HHV-7 was discovered in 1990 (12) and has also been shown to be a ubiquitous, T-cell-tropic agent capable of causing exanthem subitum (22, 34, 46). Both viruses are transmitted with high efficiency early in life, resulting in infection of most children with both viruses by the age of 3 years (4, 16, 47, 51, 54).
HHV-7 persists lifelong (like all other herpesviruses) and has been shown to establish a state of latency in peripheral blood mononuclear cells (2). HHV-7's capacity for latency and its ability to specifically infect and kill CD4+ T cells (2, 12, 13) are properties which the virus shares with the human immunodeficiency virus type 1 (HIV-1). Like HIV-1, HHV-7 binds to the CD4 molecule, and the virus uses CD4 as a component of its host cell receptor (26). However, HHV-7 does not cause progressive immunodeficiency disease and appears to establish a generally benign state of coexistence with its host. Further studies of this virus may therefore shed light not only on the mechanism whereby HHV-7 targets T lymphocytes but also on the basis for the virus' apathogenic phenotype.
Herpesviruses encode as many as 10 or more glycoproteins targeted to the virion envelope. Many of these are involved in virus attachment to the cell surface or fusion of the viral envelope with the plasma membrane (36). These viral glycoproteins may also serve as major targets for the host immune system. Information about HHV-7 glycoproteins is limited, although the virus is known to encode homologs of the glycoproteins B, H, and L found in other herpesviruses such as herpes simplex virus type 1 (HSV-1) (17, 27–29, 41, 43). HHV-7 does not, however, encode a homolog of the essential HSV-1 receptor-binding glycoprotein D (6, 23, 31, 49) or of the heparin-binding glycoprotein C (20, 21, 45).
In the present study, we have focused our attention on the analysis of a novel glycoprotein which is unique to HHV-7 and HHV-6. In HHV-6, this molecule forms a complex (gp82-105) containing several polypeptides which share extensive amino acid identity and are thought to be derived from the same gene through differential mRNA splicing (33). The gp82-105 complex is a major component of the HHV-6 virion and represents a target for virus-neutralizing antibodies (32, 33), properties consistent with a possible role in virus attachment to, or penetration of, the host cell.
The gene encoding HHV-6 gp105 spans several open reading frames (ORFs; U98 to U100) which are unique to HHV-6 and HHV-7 among the known human herpesviruses (33). We report here on the isolation, analysis, and characterization of a homologous gene in HHV-7. A 1.7-kb HHV-7 cDNA clone was identified, and a single major mRNA species of this same size (1.7 kb) was detected by Northern blot analysis of poly(A)+ RNA from HHV-7-infected cells. A polyclonal antiserum raised against the purified, recombinant protein product encoded by this cDNA reacted with a glycoprotein of approximately 65 kDa (hereafter referred to as gp65) that was present in lysates from virus-infected cells and in highly purified HHV-7 virion preparations; this glycoprotein was reduced in size to roughly 50 kDa upon digestion with N-glycanase (PNGase F). gp65-specific antiserum also neutralized the infectivity of cell-free virus inocula, providing additional support for the hypothesis that gp65 is a virion component. Finally, HHV-7 gp65 was expressed in eukaryotic cells, and its biochemical properties were examined. Gp65 was found to interact specifically with heparin and heparan sulfate proteoglycans (and not with N-desulfated heparin, N-acetylheparin, or chondroitin sulfates A and C). These findings suggest that HHV-7 gp65 may promote virus attachment to, or invasion through, cell surface proteoglycans.
MATERIALS AND METHODS
cDNA library.
The cDNA library, which was generated from HHV-7AL-infected SupT1 cells and cloned into the λZAPII vector (Stratagene, La Jolla, Calif.), has been described elsewhere (41). This library was screened with a digoxigenin-labeled (Boehringer Mannheim, Indianapolis, Ind.) DNA probe generated by PCR amplification of HHV-7 genomic DNA with oligonucleotide primers corresponding to the predicted location of the HHV-7 homolog of HHV-6 gp105 (29, 33). Primers used were as follows: outer primers, 3733 (5′-AGAACTCATTGGGAGTGCGCGGAT) and 4350 (5′-TATGTCTCTCCAGGGTTCGATGGT); and inner primers, 2737 (5′-ACTCTGTATCGTTGTTCG) and 3363 (5′-TCATCTCTTGGTCTTCTG). The HHV-7 genomic coordinates of the final DNA probe were 140,225 through 140,842 of HHV-7JI.
5′ RACE.
Total RNA was isolated from HHV-7 infected SupT1 cells by using the RNeasy Kit (Qiagen, Valencia, Calif.). The RNA was subsequently DNase treated using RQ1 RNase-free DNAse I (Promega, Madison, Wis.) for 30 min at 37°C. An aliquot of the DNase-treated RNA was analyzed on a 1.2% formaldehyde-agarose gel to assure integrity, and an additional aliquot of the DNase-treated total RNA was processed for poly(A)+ RNA by using the Oligotex Kit (Qiagen). 5′ rapid amplification of cDNA ends (RACE) was performed by using the Boehringer Mannheim 5′/3′ RACE Kit on both the total RNA and the poly(A)+ RNA templates. Briefly, single-stranded cDNA was generated from the mRNA using gene specific primer PH1 (5′-CCGTATTGGTACATATCTTTATGAG; this corresponds to nucleotides 138942 to 138966 of HHV-7JI). The cDNA was purified using the High Pure PCR Product Purification Kit (Boehringer Mannheim) and then poly(A) tailed using terminal transferase and dATP. This material was then PCR amplified using the gene specific primer PH2 (5′-TTATATAATGCAGTTGCACCAT; this corresponds to nucleotides 138978 to 138999 of HHV-7JI) and the cDNA amplification primer BMB (5′-GACCACGCGTATCGATGTCGACTTTTTTTTTTTTTTTTV; where V = A, C, or G). The PCR cycling protocol was 94°C for 2 min, followed by 10 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 40 s, followed by 25 cycles of 94°C for 15 s, 55°C for 30 s, and 72°C for 40 s (with a 20-s increase per extension cycle), followed by a final extension at 72°C. Two polymerase systems were utilized for amplification: Taq polymerase (Promega) and, for enhanced fidelity, the Expand 20kb PCR System (Boehringer Mannheim). Amplified products were analyzed on a 1.5% agarose gel and specific bands were gel isolated using the QIAquick Gel Extraction Kit (Qiagen). Gel-purified products were then cloned into the pGEM-T vector (Promega) and sequenced using the ABI PRISM DNA sequencing protocol (Perkin-Elmer, Foster City, Calif.). The sequences obtained were analyzed using the BLAST algorithm.
Northern (RNA) blot analysis.
HHV-7 infected SupT1 cells were lysed using QiASHREDDER reagent, and total poly(A)+ RNA was isolated using an Oligotex mRNA isolation kit (Qiagen). RNA quality was verified by electrophoresis through a formaldehyde-containing agarose gel, and nucleic acids were transfered to a Genescreen Plus membrane (New England Nuclear, Boston, Mass.). The resulting blot was hybridized with a radiolabeled, single-stranded, gp65 RNA probe that was generated using the T7-Riboprobe system (Promega). After overnight hybridization under conditions recommended by the manufacturer, the blot was washed thoroughly and exposed to X-ray film (Eastman Kodak) at −70°C.
Baculovirus expression of gp65.
A soluble derivative of HHV-7 gp65, bearing a carboxy-terminal polyhistidine epitope tag (His6), was expressed in insect cells by using a recombinant baculovirus expression vector. To do this, codons 23 to 468 of the gp65 cDNA were subcloned into the pMelBacB vector (Invitrogen, Carlsbad, Calif.) in frame with the honeybee mellitin signal sequence. Subcloning of gp65 sequences was achieved by PCR amplification with Pfu DNA polymerase and using the oligonucleotide primers BALA1, (5′-tcgaggatcctGAAAAAGCACGCACGGCAATAACT) and BVB1 (5′-agcgtcgacctagtggtggtggtggtggtgACCACCCATAACATTTTGTAACT) (lowercase letters denote nonviral sequences, including the translation stop codon [in boldface], while uppercase letters represent gp65-specific sequences; underlined residues are the restriction sites, and italicized residues correspond to the His6 epitope tag). After cloning into pMelBacB, recombinant baculovirus was generated using the Bac-N-Blue system (Invitrogen). PCR analysis was used to verify the presence of HHV-7 insert sequences in plaque-purified virus clones, and protein expression from these clones was verified by immunoblot analysis of culture supernatants from virus-infected High Five cells (Invitrogen) by using a His6-specific monoclonal antibody (Qiagen). For subsequent biochemical studies (e.g., heparin binding), medium containing the expressed proteins was collected, centrifuged to remove debris, and concentrated using Centricon-30 microconcentrators (Amicon, Bedford, Mass.). Protein concentrations within these samples were determined using the Bio-Rad Protein Assay Reagent.
Generation of mouse antisera to gp65.
Supernatants from High Five insect cells that were infected with a gp65-expressing baculovirus vector were collected and dialyzed extensively against 50 mM sodium phosphate–0.5 M NaCl (pH 8.0). His6-tagged gp65 was then purified using nickel-agarose (Qiagen), eluted with imidazole, and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Protein preparations containing only a single detectable band upon Coomassie blue staining of SDS-polyacrylamide gels were used for injection into a BALB/c mouse (11 μg per injection, delivered subcutaneously in Freund complete adjuvant). After initial protein injection, the animal was boosted by a second protein injection in Freund's incomplete adjuvant (29 days after the initial inoculation), and serum was collected 21 days thereafter. The specificity of this antiserum (and the corresponding prebleed serum sample) was then assessed by immunoblot analysis using His6-tagged gp65 as a target.
Generation of rabbit antisera to gp65.
The preparation of the immunogen was similar to that described above, except that the recombinant His6 tagged gp65 protein was bound to nickel-nitriloacetic acid (NTA)-agarose beads (Qiagen) which were then pelleted, washed extensively with 50 mM sodium phosphate–0.5 M NaCl–20 mM imidazole (pH 8.0), and resuspended in phosphate-buffered saline (PBS) as a 75% gel slurry. A 1-ml aliquot of this material (i.e., His6-tagged gp65 protein, bound to nickel-NTA-agarose beads) was then mixed with an equal volume of Freund's complete adjuvant and injected into a 6-month-old male New Zealand White rabbit (Cocalico Biologicals, Reamstown, Pa.). The rabbit received a boost injection in Freund's incomplete adjuvant 1 week after the initial immunization and once a month thereafter for four consecutive months. Each immunization delivered approximately 15 to 50 μg of His6-tagged HHV-7 gp65. Ten days after the last boost, the rabbit was sacrificed by exsanguination, and serum was collected. Like the mouse antiserum, this antiserum reacted specifically with gp65 upon immunoblot analysis of both purified recombinant gp65 and highly purified HHV-7 virion preparations, while the matched preimmune serum did not react with gp65 (data not shown).
Immunoblot analysis of HHV-7 virions.
A lysate of sucrose density gradient-purified HHV-7 virions (strain H7-4) was obtained from Advanced Biotechnologies, Inc. (Columbia, Md.). Aliquots of this material were then separated by SDS-PAGE, transferred to nitrocellulose, and analyzed by immunoblot analysis, using the mouse antiserum directed against gp65. The antiserum was used at a dilution of 1:100, and bound antibodies were detected using a horseradish peroxidase (HRP)-conjugated anti-mouse antibody and chemiluminescent reagents (ECL; Amersham-Pharmacia, Piscataway, N.J.). In some experiments, the viral lysate was digested with N-glycanase (Peptido-N-Glycosidase F [PNGase F]; Oxford GlycoSciences, Wakefield, Mass.) for 14 h at 37°C (2.5 U of enzyme/5 μg of viral lysate) prior to separation by SDS-PAGE and immunoblot analysis.
Radioimmunoprecipitation.
To determine whether gp65 was expressed in HHV-7-infected SupT1 cells, 6 × 106 infected or uninfected cells were radiolabeled for 18 h in 10 ml of methionine- and cysteine-free Dulbecco modified Eagle medium (Gibco, Grand Island, N.Y.) supplemented with 5% dialyzed fetal bovine serum, 2 mM glutamine, and 25 μCi of Expre35S35S-Protein Labeling Mix (New England Nuclear) per ml. After labeling, the cells were collected into pellets and lysed in 1.5 ml of PBS containing 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and 2 μg of pepstatin per ml, 7.5 μg of bestatin per ml, and 5 μg each of antipain, aprotonin, leupeptin, and soybean trypsin inhibitor per ml. After 30 min, insoluble cellular debris was removed by centrifugation. Each lysate was divided into two equal aliquots, and these were incubated with gp65-specific mouse antiserum or preimmune serum, respectively, for 2 h at 4°C. Protein A-Trisacryl beads (Pierce, Rockford, Ill.) were then added to the samples and, following a 1-h incubation, the beads were pelleted, washed with PBS–0.1% Triton X-100, and boiled in SDS-PAGE sample buffer. Eluted proteins were resolved by electrophoresis on a 12.5% polyacrylamide gel. The gel was fixed, soaked in Amplify reagent (Amersham), dried, and subjected to fluorography.
HHV-7 gp65 mutagenesis.
The 1,338-bp fragment of HHV-7 gp65 cDNA (codons 23 to 468) previously cloned in the s-pIG vector (Novagen, Madison, Wis.) was used a substrate for mutagenesis. Two putative heparin-binding domains were targeted for mutagenesis, which was performed by alanine substitution using the QuikChange mutagenesis kit (Stratagene).
Mutant clones were designated as follows: M1 (159ARHRWERR166 → 159AAAAWERR166), M2 (184FKKMRS189 → 184FAAMRS189), and M12 (both mutations introduced into the same DNA clone). The oligonucleotide primers used in the M1 and M2 mutagenesis reactions were, respectively, as follows: DM1 (5′-AGCCTGGTTTTGTTCCCTGCTgctgctgcTTGGGAAAGACGTGAGCAATA) and DM1R (5′-TATTGCTCACGTCTTTCCCAAgcagcagcAGCAGGGAACAAAACCAGGCT); and DM2 (5′-TTACAGATACGCGCAGATTTTgctgctATGCGTAGCTACAGCGGAATA) and DM2R (5′-TATTCCGCTGTAGCTACGCATagcagcAAAATCTGCGCGTATCTGTAA). The uppercase letters denote authentic HHV-7 sequences, while the lowercase letters denote mutated residues. After mutagenesis, which was conducted according to the manufacturer's protocol, a short restriction fragment spanning the entire mutated region (a 476-bp HindIII/SphI fragment; bp 353 to 820) was excised and substituted for the corresponding region within the pMelBacB-gp65-(His6) expression construct. The final clones were then sequenced to verify the presence of the desired mutations (and the lack of any additional changes), and recombinant baculovirus expression vectors encoding the mutated derivatives of HHV-7 gp65 were generated (see above).
Analysis of heparin-binding by gp65.
Culture supernatants from High Five insect cells infected with the gp65-(His6)-expressing baculovirus recombinant (10 μg of protein) were prepared in 500 μl of PBS containing 0.1% Triton X-100, in the presence or absence of 350 μg of various competitor glycosaminoglycans (heparin; heparan sulfate; chondroitin sulfates A, B, and C; N-acetylheparin; and de-N-sulfated heparin; all of these reagents were obtained from Sigma, St. Louis, Mo.). After being mixed for 1 h at 4°C, 100 μl of a 50% slurry of heparin-acrylate beads (Sigma) equilibrated in PBS–0.1% Triton X-100 (containing approximately 35 μg of immobilized heparin) was added. Samples were mixed for 2 h at 4°C, after which the beads were pelleted and unbound material was removed. The beads were subsequently washed five times with 1 ml of PBS–0.1% Triton X-100, and bound material was eluted by boiling the beads in 50 μl of SDS-PAGE sample buffer. Heparin-bound and unbound material was resolved on SDS–12.5% polyacrylamide gels and electroblotted onto nitrocellulose membranes (Amersham Pharmacia).
To detect the gp65-(His6) protein, nitrocellulose membranes were blocked overnight at 4°C in PBS–0.05% Tween 20 containing 5% nonfat dry milk; blots were probed with a Penta-His monoclonal antibody (Qiagen) in PBS–0.05% Tween 20–2% milk, followed by HRP-conjugated sheep anti-mouse immunoglobulin (Amersham Pharmacia). Bound antibodies were detected with ECL Detection Reagents (Amersham Pharmacia), and biotinylated molecular weight markers (Bio-Rad, Hercules, Calif.) were visualized by using HRP-streptavidin (Amersham Pharmacia).
Virus neutralization experiments.
Filtered (0.45 μm [pore size]) infectious supernatant obtained from HHV-7-infected SupT1 cells was used. The procedures for HHV-7 propagation in the SupT1 CD4+ lymphoblastoid T-cell line and the preparation of virus stocks have been previously described (26, 40, 42). For the virus neutralization assay, the viral inoculum was preincubated for 60 min at room temperature with rabbit anti-gp65 immune and preimmune sera. SupT1 cells were then incubated with the treated or untreated viral stocks (500 μl of virus per 250,000 cells; multiplicity of infection [MOI] of approximately 0.1) and seeded in culture in 24-well plates (Costar, Corning, N.Y.). Infection was monitored by light microscopic examination (for the detection of syncytia) and by indirect immunofluorescence staining at 48 and 72 h postinfection.
GenBank accession number.
The sequences described here have been deposited with GenBank (accession no. AF198085).
RESULTS
Identification and analysis of the HHV-7 gp65 gene.
A digoxigenin-labeled HHV-7 DNA probe corresponding to the HHV-6 gp105 gene (33) was used to screen a library of cDNAs from HHV-7AL-infected SupT1 cells, cloned into the λZAPII vector (41). After several rounds of screening, a total of six positively hybridizing clones were selected for further analysis. Plasmids were excised from the lambda phage vector and subjected to DNA sequence analysis by using oligonucleotide primers derived from the T3 and T7 promoter elements which flank the inserted cDNAs in this vector. Five of the six cDNA clones were found to contain common sequences, derived from ORFs U98 to U100 on the complementary strand of the HHV-7 genome (clones 1111, 2113, 3115, 4117, and 6119). Thus, these clones corresponded to sequences with homology to the HHV-6 gp105 gene (33). The sixth clone (clone 1116) was derived from a different gene, encoded on the opposite strand of the viral genome (27, 29). The splicing patterns and sequence contents of these various cDNAs are summarized in Table 1.
TABLE 1.
HHV-7 cDNA clones which hybridize to a HHV-6 gp105 DNA probea
| Clone | Insert size (kb) | Position (nucleotide no.)
|
Comment | |
|---|---|---|---|---|
| T3 | T7 | |||
| 1111 | 1.6 | 139006 | 137060 [poly(A)]c | gp65 (exons 2 to 8) |
| 1116 | 2.0 | 138015 | 141588b [poly(A)] | DR1 gene (Table 2) |
| 2113 | 0.9 | 137078 | 138660 | gp65 (exons 3 to 8) |
| 3115 | 0.9 | 138261 | 137057 [poly(A)] | gp65 (exons 4 to 8) |
| 4117 | 1.7 | 142655b | 136980 [poly(A)] | gp65 (intact gene) |
| 6119 | 1.4 | 141154b | 137208 [poly(A)] | gp65 (exons 2 to 8, plus novel first exon) |
All nucleotide positions refer to the coding DNA strand of the HHV-7JI genome (29).
This position maps to the DR elements at the left and right viral genomic termini; map units are provided for the location most proximal to the other elements of the cDNA (DRR). The map units for the opposite DR element (DRL) are, respectively, 2541 (clone 1116), 3608 (clone 4117), and 2107 (clone 6119).
Poly(A) indicates the location of the oligo(dT) tract in the cDNA clone. This corresponds to the putative 3′ end of the corresponding mRNA transcript (i.e., the 3′ poly(A) tail).
Complete sequence analysis of clone 1116 revealed that it corresponds to a spliced mRNA encoding the viral DR1 gene product (27, 29). As shown in Table 2, the splicing pattern detected in this cDNA was identical to that predicted by Megaw and colleagues (27), with the addition of a short upstream exon (noncoding). The major ORF contained within this cDNA clone is predicted to encode a protein of 344 amino acids. This is somewhat shorter than the DR1 gene product predicted by Megaw and coworkers, a finding presumably due to the differences in the length of the direct repeats (DRs) of HHV-7AL (which served as the source of our cDNA clones) versus the DRs of HHV-7RK (27).
TABLE 2.
Exon positions within HHV-7 cDNA clone 1116
| Exon | Position (nucleotide no.)
|
Sequencesb
|
||
|---|---|---|---|---|
| cDNA | Genomica | SA | SD | |
| 1 | 1–130 | 138015–138144 | TAG/(GT)ATGT | |
| 2 | 131–393 | 139567*–139829* | CTCTTCTATCAC(AG)/A | CAT/(GT)AAGC |
| 3 | 394–2021 [poly(A)c] | 139964*–141588* | TTGCTCTATCGC(AG)/G | |
All positions refer to the coding DNA strand of the HHV-7JI genome (29). Positions that map to the DR elements at the left and right viral genomic termini are indicated by an asterisk (map units are provided for the location most proximal to the other elements of the cDNA).
SA and SD refer to the splice acceptor and splice donor sequences, respectively, present within the HHV-7JI genome. Splice consensus motifs are indicated in parentheses. The slash indicates the position at which mRNA splicing occurs (as determined by sequence analysis of the cDNA clone).
Poly(A) indicates the location of the oligo(dT) tract in the cDNA clone. This corresponds to the putative 3′ end of the corresponding mRNA transcript (i.e., the 3′ poly(A) tail).
Since we were interested in the HHV-7 homolog of HHV-6 gp105, we directed the remainder of our attention toward the analysis and characterization of the remaining five cDNA clones. In vitro transcription and translation (IVTT) of these cDNAs, using T7 RNA polymerase, showed that a detectable protein product was generated only in the case of clone 4117 (data not shown). DNA sequencing of this clone was therefore performed. Sequence analysis using the GCG software package (8) revealed the presence of a single large ORF of 1,404 nucleotides within the cDNA. The putative initiation codon (ATG) at nucleotide 174 conformed to Kozak's rules for translational start sequences (25), having an A at position −3 and a G at position +4; this ATG was preceded by an upstream, in-frame stop codon at nucleotide 153. The ORF stop codon (TAA) was located at nucleotide 1578, and a potential polyadenylation signal (AATAAA) was detected between residues 287 and 292 on the cDNA.
The ORF contained within this cDNA clone was predicted to be capable of encoding a primary translational product of 468 amino acids (approximately 54 kDa), with a pI of 7.25, 13 cysteine residues, and 9 consensus N-linked glycosylation sites. Overall, this putative protein product exhibited 30% amino acid identity and 39% similarity to its counterpart in HHV-6, as determined by using the GAP algorithm (8). Analysis of predicted protein hydrophilicity suggested that the putative protein is likely to possess a noncleaved hydrophobic signal sequence and membrane anchor of approximately 23 amino acids, making it a tail-anchored class II membrane glycoprotein (48) like its HHV-6 counterpart.
Our predictions with respect to protein size and posttranslational modification were experimentally verified by IVTT of the cDNA insert from clone 4117. This resulted in the production of a protein of approximately 50 kD, which was converted to a molecule of roughly 65 kDa upon the addition of canine microsomes to the IVTT reaction (data not shown), suggesting that the protein is glycosylated in vivo. In light of these findings, we designated this novel viral glycoprotein gp65.
Splicing and transcription of HHV-7 gp65.
Comparison of the nucleotide sequence from the gp65-encoding cDNA with that of the HHV-7 genome revealed that the gene encoding gp65 contains eight exons, spanning almost 6 kb of the viral genome (Table 3). The first exon (nucleotides 1 to 170) maps to the viral terminal direct repeats and is predicted to be noncoding, due to the presence of an upstream, in-frame stop codon at nucleotide 153, relative to the putative initiator methionine within the cDNA (nucleotide 174). All other exons in the cDNA correspond to sequences present at the right (3′) end of the unique long segment of the viral genome. Each of the exons contained within this cDNA clone showed 100% nucleotide identity with the corresponding genomic regions from both HHV-7 strain JI and HHV-7 strain RK, emphasizing the highly conserved nature of HHV-7 genomes (27).
TABLE 3.
Exon positions within the gene that encodes gp65 in the cDNA and the HHV-7JI genome sequences
| Exon | Position (nucleotide no.)
|
Sequencesb
|
||
|---|---|---|---|---|
| cDNA | Genomica | SA | SD | |
| 1 | 1–170 | 142655*–142486* | ACG/(GT)GAGT | |
| 2 | 171–418 | 139002–138755 | AAATCTCTTCGC(AG)/A | ACA/(GT)AAGT |
| 3 | 419–700 | 138660–138379 | TTAATTCTTCTA(AG)/G | ATG/(GT)AAGC |
| 4 | 701–1057 | 138305–137949 | TACCCGCTTATT(AG)/T | AGT/(GT)AAGT |
| 5 | 1058–1135 | 137828–137751 | ATTTTTTTTTTT(AG)/A | AAT/(GT)AAGA |
| 6 | 1136–1243 | 137674–137567 | TCGTTTAGTAAC(AG)/G | CAG/(GT)AAAT |
| 7 | 1244–1357 | 137496–137383 | CTTCTTCATCCT(AG)/A | TTG/(GT)AATT |
| 8 | 1358–1676 [poly(A)c] | 137301–136980 | TTTTTTCATACC(AG)/C | |
All positions refer to the coding DNA strand of the HHV-7JI genome (29). The position that maps to the DR elements at the left and right viral genomic termini is indicated by an asterisk; map units are provided for the location most proximal to the other elements of the cDNA (i.e., DRR; map units within DRL correspond to positions 3608 to 3439).
SA and SD refer to the splice acceptor and splice donor sequences, respectively, present within the HHV-7JI genome. Splice consensus motifs are indicated in parentheses. The slash indicates the position at which mRNA splicing occurs (as determined by sequence analysis of the cDNA clone).
Poly(A) indicates the location of the oligo(dT) tract in the cDNA clone. This corresponds to the putative 3′ end of the corresponding mRNA transcript (i.e., the 3′ poly(A) tail).
As noted above, four additional cDNA clones (clones 1111, 2113, 3111, and 6119) also contained fragments of the U98 through U100 viral ORFs (Table 1). Clone 1111 was found to possess the same predicted coding capacity as clone 4117 (i.e., the same ATG and TAA codons at nucleotide positions 138,998 and 137,080 of the HHV-7JI genome, respectively), while clones 2113 and 3115 were determined to contain exons 3 to 8 and 4 to 8 of the gp65 gene, respectively. These clones may therefore represent incomplete cDNA clones that lack an intact 5′ end (Table 1). Finally, clone 6119 had a very similar organization to that of clone 4117 and also spanned exons 2 to 8 of the gp65 gene, terminating at an internal oligonucleotide A stretch within exon 8. The first in-frame ATG codon in this clone was the same as that found in clone 4117. However, exon 1 of clone 6119 was derived from a different region of the viral direct repeats (nucleotide positions 141,154 to 141,070 of the HHV-7JI genome) compared to exon 1 of clone 4117. Sequence analysis of this clone suggested that this new upstream region did not change the predicted translation product of the cDNA relative to clone 4117.
Since the 5′ ends of the gp65-encoding cDNA clones 4117 and 6119 were different, we undertook to map the 5′ end(s) of the gp65-encoding mRNA(s) by using 5′-RACE analysis. In order to do this, we generated oligonucleotide primers corresponding to the 5′ end of exon 2 of the cDNA clones 4117 and 6119 (this region is conserved in both of these cDNAs; see Fig. 1). We then used this primer to perform PCR amplification of cDNA fragments corresponding to the 5′ end(s) of the gp65 mRNA(s); the PCR-derived cDNA fragments were then cloned and subjected to sequence analysis.
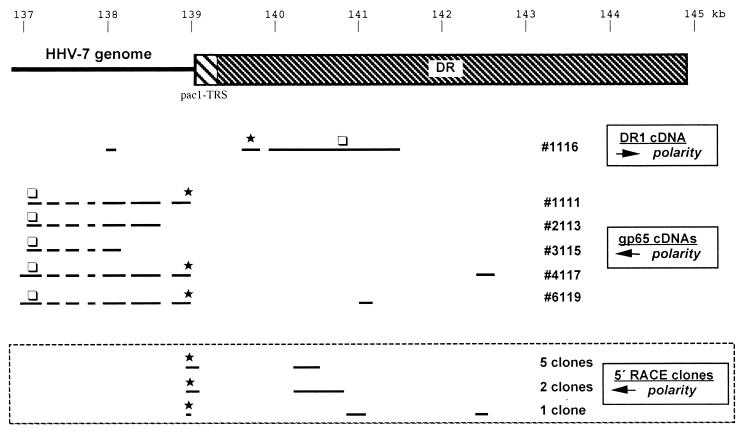
FIG. 1.
Schematic summary of cDNA clones and 5′-RACE clones analyzed in this study. The upper portion of this figure shows a schematic representation of the right end of the HHV-7 genome, including the DR and the pac1 and TRSs located at the boundary between the DR and the right end of the unique portion of the viral genome. All genomic positions are relative to the JI strain of HHV-7 (in kilobases). The lower portion of the figure shows the structure of the various cDNA and 5′-RACE clones which are described in this study. These include cDNA clone 1116, which corresponds to a putative DR1 cDNA, as well as five cDNAs (intact and partial) which correspond to putative gp65-encoding cDNAs. In all cases, the location of the first methionine within the major ORF of each cDNA is indicated by an asterisk, while the location of the stop codon is located by an open box. Note that the DR1 cDNA is of opposite polarity to the gp65 cDNAs. Finally, the structures of the 5′-RACE clones are shown in the box at the bottom of the figure. A total of eight RACE clones were sequenced, and their structures were found to fall into three groups (as shown). Seven of the eight clones were found to extend into the HHV-7 DR before undergoing a splicing event. As a consequence, these clones all contained the pac1 motif and a short array of telomeric repeat sequences. One RACE clone lacked the pac1-TRS elements but contained portions of the exon 1 from cDNA clone 6119, as well as portions of exon 1 from cDNA clone 4117. Importantly, all of the RACE clones contained an in-frame stop codon upstream of the first methionine codon (the stop codon is not marked on this figure, but the methionine codon is denoted by an asterisk). Thus, the predicted protein-coding capacity of these RACE-derived clones appears to be identical to that of gp65 cDNA clone 4117.
The sequencing results revealed a somewhat unexpected pattern of RNA 5′ ends (see Table 4). In no case did we generate clones with structures identical to clones 4117 or 6119. Rather, seven of eight clones analyzed were found to extend into the HHV-7 DR before undergoing a splicing event (Table 4). As a consequence, these clones all contained the pac1 motif and a short array of telomeric repeat sequences (TRSs) derived from the 5′ end of the right DR. This splicing pattern may have been missed in the cDNA clones due to the instability of the viral TRS motifs in Escherichia coli (unpublished data). One additional RACE clone lacked the pac1-TRS elements but contained portions of exon 1 from clone 6119, as well as portions of exon 1 from clone 4117 (Table 4).
TABLE 4.
Exon positions within 5′-RACE clones derived from gp65 mRNAs
| No. of clones | Exon | Genomic nucleotide positiona | Sequencesb
|
|
|---|---|---|---|---|
| SA | SD | |||
| 2 | 1 | 140839*–104359* | CGA/(GT)GTTT | |
| 2 | ∼139140*†(−138978) | Not defined | ||
| 5 | 1 | 140632*–104359* | CGA/(GT)GTTT | |
| 2 | ∼139140*†(−138978) | Not defined | ||
| 1 | 1 | 142603*–142486* | ACT/(GT)GAGT | |
| 2 | 141106*–140904* | GGCACTGTCGAT(AG)/G | CGG/(GT)GTGT | |
| 3 | 139002*(−138978) | AAATCTCTTCGC(AG)/A | ||
All positions refer to the coding DNA strand of the HHV-7JI genome (29). Note that all of the 5′-RACE clones have a 3′ end that is defined by the primer used in the PCR amplification reaction (this primer ends at HHV-7JI genomic nucleotide position 138978). ∗, Position maps to the DR elements at the left and right viral genomic termini (map units are provided for the location most proximal to the other elements of the cDNA [i.e., DRR]); †, position could not be accurately defined because the cDNA sequences map to a region of the viral DR that contains multiple degenerate copies of telomeric repeat sequence motifs.
SA and SD refer to the splice acceptor and splice donor sequences, respectively, present within the HHV-7JI genome. Splice consensus motifs are indicated in parentheses. The slash indicates the position at which mRNA splicing occurs (as determined by sequence analysis of the cDNA clone).
These findings confirmed the heterogeneous nature of the transcripts encoding HHV-7 gp65, at least in terms of their 5′ ends (this information is summarized in schematic form in Fig. 1). However, all eight of the 5′-RACE clones were found to contain an in-frame stop codon located upstream of the putative initiator methionine codon identified in cDNA clones 4117 and 6119. Thus, the expected protein-coding capacity of the RACE-derived cDNA clones was found to be identical to that of gp65 cDNA clone 4117.
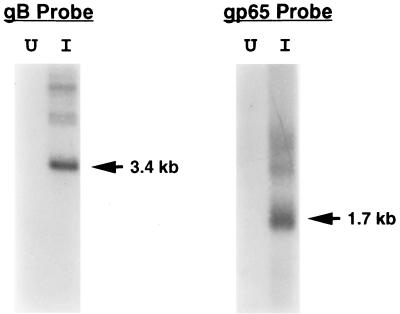
Northern (RNA) blot analysis of poly(A)+ RNA, using a single-stranded radiolabeled RNA probe generated from plasmid clone 4117, revealed the presence of a single major mRNA species of approximately 1.7 kb in poly(A)+ RNA from virus-infected SupT1 cells but not in control (virus-negative) SupT1 cells (Fig. 2). Larger, but less-abundant RNA species were also detected, which may represent splicing intermediates or the products of differential mRNA splicing, as has been suggested for the HHV-6 gp105 gene (33). Hybridization of the same RNA preparations with a double-stranded radiolabeled DNA probe corresponding to the HHV-7 glycoprotein B gene (43) resulted in the detection of a single major hybridizing species of roughly 3.4 kb (Fig. 2). This finding is consistent with a previously published report (41). The detection of higher-molecular-weight hybridization signals with the gB probe (Fig. 2) has also been described and was previously attributed to larger RNA precursors (41).
FIG. 2.
Northern blot analysis of HHV-7 gp65. Poly(A)+ RNA was isolated from HHV-7-infected SupT1 cells (I) or from uninfected cells (U), and equal amounts of RNA were then subjected to denaturing agarose gel electrophoresis and transferred to a nylon membrane. The blot was hybridized with a single-stranded, radiolabeled RNA probe corresponding to HHV-7 gp65 or to a radiolabeled DNA probe corresponding to HHV-7 gB (as indicated) and exposed to X-ray film. Shown is a photograph of the resulting autoradiogram. The arrows denote the major RNAs which hybridized to the probes used; sizes were estimated on the basis of the mobility of RNA molecular weight markers (Life Technologies, Inc.).
Biochemical analyses of HHV-7 gp65.
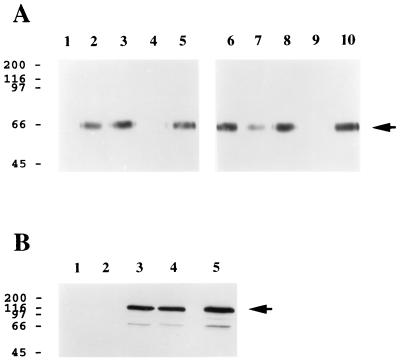
Inspection of the predicted amino acid sequence of gp65 revealed the presence of two putative heparin-binding Cardin-Weintraub consensus motifs (7): (i) at amino acid 159, ARHRWERR (this motif is of the type XBBBXXBX, where B is a basic amino acid and X represents any amino acid); and (ii) at amino acid 184, FKKMRS (this is of the type XBBXBX). In light of the presence of these sequence motifs, we theorized that gp65 might be capable of binding to cellular glycosaminoglycans. To test this hypothesis, we generated a recombinant baculovirus expression vector which encoded a soluble derivative of HHV-7 gp65 bearing a carboxy-terminal histidine tag (His6) that could be used to facilitate protein detection and purification. Culture supernatants from insect cells infected with this baculovirus vector were reacted with heparin-acrylate beads, in the presence or absence of an excess of various glycosaminoglycans. After an extensive washing, the beads were boiled in SDS-PAGE sample buffer, and the eluted gp65-(His6) was detected by immunoblot analysis using the His6-specific monoclonal antibody. Representative results are presented in Fig. 3A.
FIG. 3.
HHV-7 gp65 exhibits heparin-binding activity. (A) Insect cell culture supernatants containing soluble recombinant gp65-(His6) were incubated with heparin-acrylate beads in the absence (lanes 5 and 10; negative control) or presence of an excess amount of various glycosaminoglycans, including heparin (lanes 4 and 9; positive control), heparan sulfate (lane 1), N-acetylheparin (lane 2), de-N-sulfated heparin (lane 3), and the chondroitin sulfates A, B, and C (lanes 6, 7, and 8, respectively). After a washing, the beads were boiled in SDS-PAGE sample buffer, and eluted proteins were separated by electrophoresis on an SDS–12.5% polyacrylamide gel and transferred to nitrocellulose. Immunoblot analysis was then conducted using an anti-His6 monoclonal antibody and a chemiluminescent detection system (see Materials and Methods). (B) Lysates from COS-1 cells that were transiently transfected with a plasmid expression construct encoding lacZ-(His6) were incubated with heparin-acrylate beads in the absence (lanes 2 and 4) or presence (lanes 1 and 3) of heparin. Heparin-acrylate-bound material (lanes 1 and 2) and unbound material (lanes 3 and 4) were then subjected to immunoblot assay using an anti-His4 monoclonal antibody. Lane 5 represents a positive control in which the transfected cell lysate was directly submitted to immunoblot analysis without prior treatment with heparin-acrylate beads. In both panels, the numbers on the left represent molecular mass markers (in kilodaltons), while the arrows denote gp65-(His6) (A) and lacZ-(His6) (B), respectively. The experiments were repeated three times (A) or twice (B) with similar results.
The data show that gp65-(His6) bound to heparin-acrylate beads (Fig. 3A, lanes 5 and 10) and that this binding was competitively inhibited by the addition of excess soluble heparin to the reaction mixture (Fig. 3A, lanes 4 and 9). Heparan sulfate was also found to be an effective competitor for gp65 binding to heparin-acrylate beads (Fig. 3A, lane 1), while chondroitin sulfates A and C failed to inhibit the interaction of gp65 with heparin-coated beads (Fig. 3A, lanes 6 and 8), as did chemically modified heparin derivatives which lack the N-sulfate group (N-acetylheparin and de-N-sulfated heparin; Fig. 3A, lanes 2 and 3, respectively).
Previous studies have noted that histidine, while basic, is not enriched in heparin-binding peptide sequences (5). Furthermore, long stretches of basic amino acids are uncommon in heparin-binding proteins (14), and histidine itself appears to be a rather poor heparin binder (5, 14). Nonetheless, we performed additional control studies to verify experimentally that the His6 epitope present in our recombinant HHV-7 gp65 did not bind to heparin. To do this, cells were transfected with an expression vector encoding a recombinant derivative of E. coli β-galactosidase (LacZ) that contains an C-terminal His6 epitope tag (pcDNA3.1MycHisLacZ+; Invitrogen). Lysates from these transfected cells were then reacted with heparin-acrylate beads, and the bound (Fig. 3B, lanes 1 and 2) or unbound (Fig. 3B, lanes 3 and 4) fractions were analyzed by immunoblot analysis using a His4-specific monoclonal antibody; binding experiments were conducted in the presence (Fig. 3B, lanes 1 and 3) or absence (Fig. 3B, lanes 2 and 4) of excess soluble heparin. As is evident from the data presented in Fig. 3B, the presence of the C-terminal His6 epitope tag in E. coli lacZ did not confer the ability to bind to heparin on the protein. Furthermore, radiolabeled gp65 produced in baculovirus without the histidine tag was also shown to bind to heparin-acrylate beads (data not shown).
Having concluded that gp65-(His6) interacts specifically with heparin but not with several other closely related glycosaminoglycans, we proceeded with experiments designed to define the regions within gp65 that might contribute to heparin binding. First, short biotinylated peptides were synthesized (ADFKKMRSYS and PARHRWERRE) that corresponded to the two putative heparin-binding motifs within HHV-7 gp65 (residues 182 to 191 and residues 158 to 167, respectively). The two HHV-7 peptides both bound to radiolabeled heparin, while an irrelevant peptide from adenovirus type 7 fiber protein (GSFNPVYP) did not do so (data not shown). Furthermore, this binding could be inhibited by the addition of excess cold heparin but not by the addition of N-acetylheparin or de-N-sulfated heparin, suggesting that the binding was specific (data not shown).
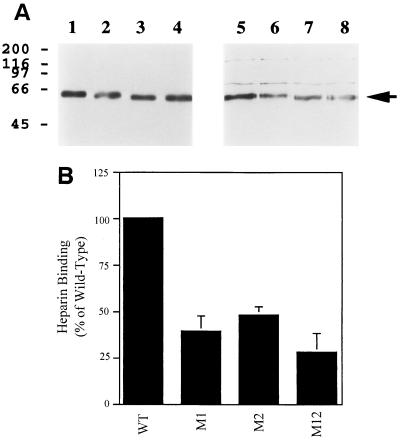
In order to confirm that the putative heparin-binding domains within gp65 were functional in the context of the intact protein, site-directed mutagenesis studies were conducted. Three mutants of gp65 were constructed: (i) M1 (159ARHRWERR166 → 159AAAAWERR166), (ii) M2 (184FKKMRS189 → 184FAAMRS189), and (iii) M12 (which contains both of these mutations). These mutants were expressed with a C-terminal (His6) epitope tag in insect cells, using the baculovirus system, and tested for their ability to bind to heparin-acrylate beads. As shown in Fig. 4, each of the individual gp65 mutants exhibited decreased binding to heparin (binding was approximately 40 to 50% of the wild-type level in both cases; Fig. 4B). The double mutant, which lacks both of the putative heparin-binding motifs, exhibited an even lower level of binding (approximately 28% of the wild-type level; Fig. 4B). In all cases, binding was competed away entirely in the presence of an excess of soluble heparin (data not shown).
FIG. 4.
Analysis of heparin-binding by mutated derivatives of HHV-7 gp65. Mutated derivatives of HHV-7 gp65 were constructed, and the proteins were then expressed in insect cells, with a C-terminal (His6) epitope tag. Heparin-binding by the proteins was then assessed (see Fig. 3 for methods). (A) The results of one representative heparin-binding experiment are shown. Lanes are as follows: 1 to 4, input protein (directly loaded onto the gel and then probed by immunoblotting with the anti-His6 monoclonal antibody); 5 to 8, protein bound to heparin-acrylate beads and then analyzed by immunoblotting with the anti-His6 monoclonal antibody. Wild-type gp65-(His6) was analyzed in lanes 1 and 5, while mutant derivatives of gp65-(His6) were analyzed in the other lanes (lanes 2 and 6, M1; lanes 3 and 7, M2; lanes 4 and 8, M12). (B) Results from four independent heparin-binding experiments (including the one shown in panel A) were densitometrically quantitated and normalized in terms of input protein loaded (as detected by the anti-His6 immunoblot assay). The amount of the mutant proteins which bound to heparin-agarose beads was then expressed as a percentage of heparin-binding by wild-type gp65-(His6), and mean binding values were plotted in the figure; bars represent the standard error of the means.
Taken together, the data presented in Fig. 4 show that each of the two putative heparin-binding domains within HHV-7 gp65 contributes to the ability of the intact protein to bind to heparin. However, it also appears that other protein domains may contribute to heparin-binding, since the double mutant still binds detectably to heparin. Additional studies will be required to define these additional heparin-binding regions within gp65.
HHV-7 gp65 is expressed in virus-infected cells and is present within highly purified virion preparations.
The results of the heparin-binding experiments suggested the possibility that HHV-7 gp65 might play a role in the process of virus binding or entry into host cells. This would be consistent with the fact that soluble heparin can block HHV-7 infection (43). We therefore decided to examine whether gp65 was present in HHV-7-infected cells and in purified virion preparations derived from such cells. To do this, a polyclonal mouse antiserum was raised against the baculovirus-derived gp65-(His6) protein. The specificity of this antiserum was then verified by performing enzyme-linked immunosorbent assay and immunoblot analyses using purified recombinant gp65-(His6) (data not shown).
The immune serum was used to conduct immunoprecipitation analyses of radiolabeled lysates from HHV-7-infected SupT1 cells, and control (uninfected) SupT1 cells. As shown in Fig. 5, the antiserum reacted specifically with a single major protein species of approximately 65 kD in the virus-infected cell lysates. The immune serum did not react with lysates from uninfected SupT1 cells (Fig. 5), and the matched preimmune serum failed to react with lysates from either virus-infected or uninfected SupT1 cells (data not shown). The reactivity of the gp65-specific mouse antiserum with the recombinant gp65-immunoglobulin (gp65-Ig) fusion protein is shown in Fig. 6A. As expected, the corresponding preimmune serum failed to react with this recombinant gp65-Ig fusion protein (Fig. 6A).
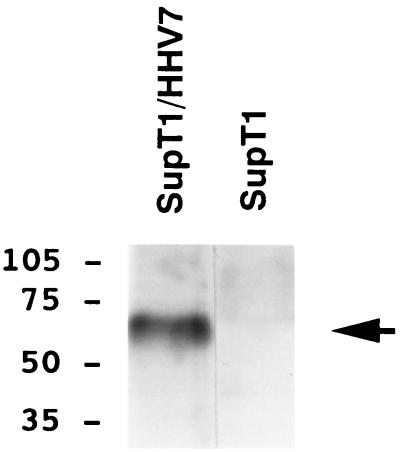
FIG. 5.
HHV-7 gp65 is expressed in HHV-7-infected cells. A murine polyclonal antiserum was raised against the baculovirus derived gp65-(His6) protein and used to perform immunoprecipitation analysis of radiolabeled cell lysates from HHV-7-infected SupT1 cells (SupT1/HHV7) and from uninfected SupT1 cells (SupT1). The immune antiserum reacted with a single major protein species of approximately 65 kDa (indicated by the arrow). The preimmune serum did not react with this protein (data not shown; see also Fig. 6).
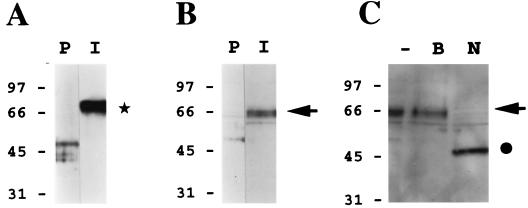
FIG. 6.
HHV-7 gp65 is present within highly purified HHV-7 virion preparations. A murine gp65-specific polyclonal antiserum (see Fig. 5) was used to perform immunoblot analyses of sucrose density gradient-purified HHV-7 virion preparations. (A) The immune antiserum (I) or its preimmune counterpart (P) was reacted with a purified gp65-Ig fusion protein (this fusion protein contained amino acid residues 23 to 325 of gp65, fused in frame to the Fc portion of human immunoglobulin G, contained in the signal pIg-Tail vector; Novagen, Inc.). As shown by the asterisk, only the immune antiserum reacted with the gp65-Ig protein (ca. 70 kDa). (B) The immune antiserum (I) or its preimmune counterpart (P) was reacted with a lysate of sucrose density gradient-purified HHV-7 virions. The arrow denotes the specific reactivity of the immune antiserum with a protein doublet of roughly 65 kDa. (C) The HHV-7 virion lysate was subjected to extensive digestion with N-glycanase (PNGase F) prior to immunoblot analysis with the immune antiserum (N). In control experiments, the lysate was incubated in reaction buffer alone (i.e., no PNGase was added) under the same conditions (B), or the lysate was analyzed directly, without prior treatment in vitro (−). The arrow denotes the position of HHV-7 gp65, while the filled circle shows the size of the product of PNGase digestion of this molecule (ca. 50 kDa).
The gp65-specific antiserum was next used to perform an immunoblot analysis of a lysate of sucrose density gradient-purified HHV-7 virions. As shown in Fig. 6B, the immune antiserum reacted with a protein doublet of approximately 65 kDa in the virion preparation (denoted by the arrow), which was not detected by the preimmune serum. Finally, to confirm that gp65 was glycosylated, we conducted in vitro deglycosylation experiments using N-glycanase (PNGase F). As shown in Fig. 6C, enzymatic deglycosylation of the HHV-7 virion preparation reduced gp65 to a putative precursor form of roughly 50 kDa (denoted by the filled circle).
The findings shown in Fig. 6 strongly suggest that gp65 is a component of HHV-7 virions. However, further analysis revealed that the highly purified virion preparation contained not only viral proteins (gp65) but also cellular membrane components (e.g., transferrin receptor and CD3; data not shown). This raised the possibility that viral nonstructural proteins might also be present within the these virion preparations. We therefore performed additional experiments to confirm that gp65 is indeed a component of present in HHV-7 virions.
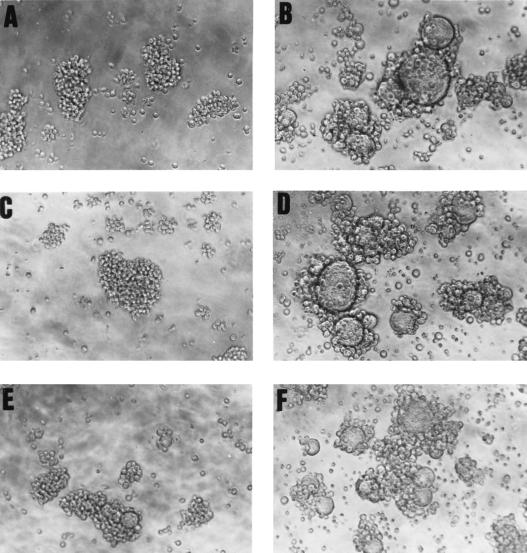
Immunoelectron microscopy studies using the mouse polyclonal antiserum have been unsuccessful to date, and we therefore resorted to more-indirect experiments, designed to test if gp65-specific antisera might neutralize the infectivity of HHV-7. These studies could not be performed using the gp65-specific mouse antiserum, since they required relatively large volumes of serum. Thus, it was necessary to generate a polyclonal rabbit antiserum directed against HHV-7 gp65 using the recombinant baculovirus-derived gp65-(His6) immunogen (see Materials and Methods). After verifying that this serum reacted specifically with recombinant forms of gp65 [gp65-Ig and gp65-(His6)] and with native gp65 from virion preparations (data not shown), we then used it to conduct virus neutralization experiments, using high-titer, cell-free virus inocula. Results from a representative neutralization experiment are shown in Fig. 7. It can be readily appreciated that the gp65-specific immune antiserum efficiently blocked virus-induced cytopathic effects (syncytium formation) in SupT1 cells (Fig. 7C and E), whereas the corresponding preimmune serum had no effect on virus-induced syncytium formation (Fig. 7D and E). This finding was corroborated by immunofluorescence analysis of viral antigen expression in these cultures (data not shown) and was confirmed in a replicate experiment. Taken together with the data in Fig. 6, these findings strongly suggest that HHV-7 gp65 is indeed a virion glycoprotein, with a molecular size that is consistent with the predicted size of the product of our cloned cDNA species.
FIG. 7.
A gp65-specific antiserum neutralizes virus infectivity. An infectious, cell-free stock of HHV-7 was generated, and preincubated for 60 min at room temperature with a diluted aliquot of rabbit polyclonal antiserum raised against the baculovirus-derived gp65-(His6) protein. SupT1 cells were then incubated with the treated virus preparation (MOI of ∼0.1), and infection was monitored by light microscopic examination of syncytium formation at 72 h following virus infection. Panels: A, negative control (uninfected SupT1 cells, with no added virus); B, positive control (SupT1 cells infected with HHV-7 in the absence of any antiserum); C and E, SupT1 cells exposed to the virus-containing inoculum and preincubated with gp65-reactive immune serum at a dilution of either 1:25 or 1:100, respectively; D and F, SupT1 cells exposed to the virus-containing inoculum and preincubated with preimmune rabbit serum at a dilution of either 1:25 or 1:100, respectively. Note the presence of extensive syncytia in panels B, D, and F, but not in the panels in which the virus inoculum was treated with the gp65-specific serum (panels C and E). Magnification, ×250.
DISCUSSION
Foa-Tomasi and colleagues reported that seven glycoproteins were immunoprecipitated from HHV-7-infected cells by human serum antibodies, with molecular masses of 100, 89, 82, 67, 63, 53, and 43 kDa (11). While the genes which encode these glycoproteins have not been determined, it is likely that at least some of them may correspond to glycoproteins B, H, and L. These glycoproteins have been found to have molecular masses of roughly 100 to 110 kDa (17, 43), 80 to 90 kDa (28, 41), and 33 to 37 kDa (28), respectively. Thus, either or both of the 67- and 63-kDa glycoproteins previously identified by Foa-Tomasi and coworkers could conceivably represent products of the gp65 gene (ORFs U98 to U100). This would be consistent with the size of the virion form of gp65 which we detected here. Further studies will be required to determine if this is indeed the case. However, we have been able to show that antibodies present in sera from HHV-7-positive children recognized Ni-agarose-purified gp65-(His6) in an immunoblot assay, while the protein was not detected by sera from HHV-7-negative children (data not shown).
The predicted size of HHV-7 gp65, at 468 amino acids, is considerably shorter than its counterpart in HHV-6 (650 amino acids) (33). This difference is consistent with the compressed structure of the right (3′) end of the unique long segment of the HHV-7 genome compared to that of HHV-6 (15, 27, 29). Furthermore, two ORFs which contribute to the HHV-6 gp105 gene (ORFs 96 and 97) and which map to this genomic region are lacking in HHV-7.
Like HHV-6 gp105, HHV-7 gp65 is encoded by a highly spliced mRNA. Splice donor and acceptor consensus sequences were identified in the viral genome at the boundaries of each of the exons (Table 3), and the overall pattern of splicing was very similar to that predicted by Megaw and colleagues, based on an analysis of the HHV-7 genome. These authors suggested that the gp65 gene from HHV-7RK, which they termed ORF U100, contain 10 exons (27). The first seven of these exons were found in the present gp65 cDNA (clone 4117, Table 3). However, two differences were observed between Megaw's prediction and our experimentally determined findings. First, the putative splice donor at the end of exon 7 was not used in this cDNA, which therefore lacks the final three downstream exons (8 to 10) that were predicted by Megaw et al. (27). Second, an unanticipated, noncoding, upstream exon was found in our gp65 cDNA, which was derived from a position within the viral DRs. This suggests that the promoter element for this gene may be located within the DRs, a finding that was supported by our 5′-RACE analysis. It also raises the possibility that the gene could be expressed from the DRL element, following genome circularization within the infected cell.
Our Northern blot analyses revealed that the major gp65 RNA species present in virus-infected cells was approximately 1.7 kb in length (i.e., the same size as our cDNA clone). This suggests that the cDNA clone obtained is full length. This interpretation is reinforced by the fact that the apparent molecular size of the in vitro-translated protein product of the cDNA clone (∼50 kDa) was very similar to the observed size of the enzymatically deglycosylated form of virion gp65 (also ∼50 kDa). The fact that our gp65-specific antiserum reacted with two distinct gp65 isoforms of roughly 65 kDa in virion preparations may suggest some variation in the posttranslation modification of gp65, since only a single protein species was detected after PNGase treatment.
Predictive analysis of the gp65 sequence revealed the presence of two putative heparin-binding motifs and prompted us to speculate that gp65 might represent a heparin-binding glycoprotein. This hypothesis was experimentally validated by using a soluble recombinant derivative of gp65. This glycoprotein was found to interact specifically with heparin and heparan sulfate proteoglycans but not with de-N-sulfated heparin or chondroitin sulfates A and C. The selectivity of gp65 for heparin and heparan sulfate, but not for chondroitin sulfates A and C, strongly suggests that this binding interaction is specific for heparan sulfate proteoglycans (37, 44). This interpretation is supported by the fact that gp65 binding to heparin was dependent on N-sulfation, since neither N-acetylheparin nor de-N-sulfated heparin could competitively inhibit gp65 binding to heparin-acrylate beads.
The functional relevance of the two putative heparin-binding sites within HHV-7 gp65 was confirmed by using two complementary approaches. First, short peptides corresponding to the two domains were synthesized and examined for their ability to bind to radiolabeled heparin. Second, site-directed mutations were introduced into the intact gp65 protein at each of the putative heparin-binding motifs (individually or in combination). Heparin-binding by these full-length mutant derivatives of gp65 was then examined. The two assay systems yielded similar and consistent findings. Each of the two domains examined was found to be capable of binding to heparin in a functional assay, and both domains were found to contribute to heparin binding by the intact protein. The data also suggest that at least one more region must contribute to heparin binding by HHV-7 gp65, since a mutated version of the protein that lacks both of the predicted binding sites was found to retain the ability to bind to heparin (albeit with reduced efficiency). Precedent for this exists, in that the pseudorabies virus glycoprotein C has been shown to contain multiple discrete units that can function independently of one another to mediate binding to heparan sulfate proteoglycans (HSPGs) (10).
In addition to gp65, HHV-7 also encodes a second glycoprotein which has been found to bind to cell surface heparan sulfate proteoglycans (glycoprotein B), and soluble heparin has been shown to inhibit viral infection and syncytium formation in the SupT1 T-cell line (43). The presence of two heparin-binding glycoproteins within a single virus is not unexpected. Indeed, all well-studied human alpha- and betaherpesviruses contain at least two glycoproteins which bind to HSPGs: a glycoprotein B homolog (found in all alpha- and betaherpesviruses [reviewed in reference 30]) and a member of either the gC family (for alphaviruses [19, 21]) or gC-II (for human cytomegalovirus [24]). Presumably, the presence of two heparin-binding glycoproteins within the same virus indicates the importance of cell surface proteoglycans as receptors for viral attachment (reviewed in reference 37). In addition, it is possible that HHV-7 gB and gp65 may exhibit differences in their relative affinity for distinct cell surface heparan sulfate proteoglycans, as do HSV-1 gB and gC (18).
It is intriguing to note that HHV-6A (GS) gp105 also contains three putative heparin-binding motifs of the type XBBXBX (94LKRVKA99, 197MRRLKP202, and 619PRKLRC624), whereas the predicted protein products of two recently identified HHV-6B (Z29) gp105 cDNAs (B. Chandran, unpublished data) contain only a single consensus heparin binding motif of the type XBBXBX (291VHHDRP296 in the Z29 U100 ORF from the complete genome of HHV-6B strain Z29). The single putative heparin-binding motif within HHV-6B gp105 is likely to be weak at best, since histidine, while basic, is not enriched in heparin-binding peptide sequences (5) and appears to be a rather poor heparin binder (5, 14). Thus, it is conceivable that differences in the interaction of HHV-6 gp105 glycoproteins with cell surface heparan sulfate proteoglycans could contribute to differences in the observed host cell tropism of HHV-6A and HHV-6B. Additional experiments will be required to test this hypothesis.
In conclusion, the findings reported here suggest that HHV-7 encodes two heparin-binding glycoproteins which may play a role in the process of virus infection (glycoprotein B and gp65 [identified for the first time here]). These molecules may contribute to the virus' ability to attach to and enter human cells, including salivary gland cells (3, 9, 50, 52) and primary human CD4+ T lymphocytes (12). Further studies of HHV-7 and its encoded glycoproteins may shed light on the biological properties of this ubiquitous human virus and may provide insights that could contribute to the development of gene delivery vehicles capable of targeting CD4+ T cells (12) and/or the salivary gland (1, 39).
ACKNOWLEDGMENTS
We thank Caroline Hall and Renee Norton for providing HHV-7 stocks and virus-infected SupT1 cells, John Frelinger and Rick Willis for assistance with the generation of mouse antisera, Robert Rose and Chris Lane for assistance with baculovirus propagation and preparation, George Kampo and Jack Maniloff for preparation of oligonucleotides and for performing DNA sequence analyses, and Bo Wisdom (Kansas University Medical Center Protein Core Facility) for preparation and purification of synthetic peptides. We also thank Robert Linhardt and Ishan Capila for assistance with the identification of putative heparin-binding elements within HHV-7 gp65 and Greg Williams and James Whitman of Advanced Biotechnologies, Inc., for expert advice and assistance.
This work was supported by NIH grants to S.D. (R21 AI34231 and KO4 AI01240), by NIH Training Grants T32 CA09363 and T32 AI07362 to D.S. and P.H., and by Telethon grant 279/bi to P.S.
REFERENCES
- 1.Baum B. Salivary gland repair. Adv Dental Res. 1995;9:22. doi: 10.1177/0895937495009003S1301. [DOI] [PubMed] [Google Scholar]
- 2.Berneman Z N, Ablashi D V, Li G, Eger-Fletcher M, Reitz M S, Jr, Hung C L, Brus I, Komaroff A L, Gallo R C. Human herpesvirus 7 is a T-lymphotropic virus and is related to, but significantly different from, human herpesvirus 6 and human cytomegalovirus. Proc Natl Acad Sci USA. 1992;89:10552–10556. doi: 10.1073/pnas.89.21.10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black J B, Inoue N, Kite-Powell K, Zaki S, Pellett P E. Frequent isolation of human herpesvirus 7 from saliva. Virus Res. 1993;29:91–98. doi: 10.1016/0168-1702(93)90128-a. [DOI] [PubMed] [Google Scholar]
- 4.Black J B, Schwarz T F, Patton J L, Kite-Powell K, Pellett P E, Wiersbitzky S, Bruns R, Muller C, Jager G, Stewart J A. Evaluation of immunoassays for detection of antibodies to human herpesvirus 7. Clin Diagn Lab Immunol. 1996;3:79–83. doi: 10.1128/cdli.3.1.79-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell E E, Nadkarni V D, Fromm J R, Linhardt R J, Weiler J M. Importance of specific amino acids in protein binding sites for heparin and heparan sulfate. Int J Biochem Cell Biol. 1996;28:203–216. doi: 10.1016/1357-2725(95)00123-9. [DOI] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume G, Arsenakis M, Farabegoli F, Roizman B. Entry of herpes simplex virus 1 in BJ cells that constitutively express viral glycoprotein D is by endocytosis and results in degradation of the virus. J Virol. 1988;62:159–67. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardin A D, Weintraub H J. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 8.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Luca D, Mirandola P, Ravaioli T, Dolcetti R, Frigatti A, Bovenzi P, Sighinolfi L, Monini P, Cassai E. Human herpesviruses 6 and 7 in salivary glands and shedding in saliva of healthy and human immunodeficiency virus positive individuals. J Med Virol. 1995;45:462–468. doi: 10.1002/jmv.1890450418. [DOI] [PubMed] [Google Scholar]
- 10.Flynn S J, Ryan P. The receptor-binding domain of pseudorabies virus glycoprotein gC is composed of multiple discrete units that are functionally redundant. J Virol. 1996;70:1355–1364. doi: 10.1128/jvi.70.3.1355-1364.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foa-Tomasi L, Avitabile E, Ke L, Campadelli-Fiume G. Polyvalent and monoclonal antibodies identify major immunogenic proteins specific for human herpesvirus 7-infected cells and have weak cross-reactivity with human herpesvirus 6. J Gen Virol. 1994;75:2719–2727. doi: 10.1099/0022-1317-75-10-2719. [DOI] [PubMed] [Google Scholar]
- 12.Frenkel N, Schirmer E C, Wyatt L S, Katsafanas G, Roffman E, Danovich R M, June C H. Isolation of a new herpesvirus from human CD4+ T cells. Proc Natl Acad Sci USA. 1990;87:748–752. doi: 10.1073/pnas.87.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frenkel N, Wyatt L S. HHV-6 and HHV-7 as exogenous agents in human lymphocytes. Dev Biol Stand. 1992;76:259–265. [PubMed] [Google Scholar]
- 14.Fromm J R, Hileman R E, Caldwell E E, Weiler J M, Linhardt R J. Pattern and spacing of basic amino acids in heparin binding sites. Arch Biochem Biophys. 1997;343:92–100. doi: 10.1006/abbi.1997.0147. [DOI] [PubMed] [Google Scholar]
- 15.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 16.Hall C B, Long C E, Schnabel K C, Caserta M T, McIntyre K M, Costanzo M A, Knott A, Dewhurst S, Insel R A, Epstein L G. Human herpesvirus-6 infection in children. A prospective study of complications and reactivation. N Engl J Med. 1994;331:432–438. doi: 10.1056/NEJM199408183310703. [DOI] [PubMed] [Google Scholar]
- 17.Hata A, Mukai T, Isegawa Y, Yamanishi K. Identification and analyses of glycoprotein B of human herpesvirus 7. Virus Res. 1996;46:125–137. doi: 10.1016/s0168-1702(96)01395-0. [DOI] [PubMed] [Google Scholar]
- 18.Herold B C, Gerber S I, Belval B J, Siston A M, Shulman N. Differences in the susceptibility of herpes simplex virus types 1 and 2 to modified heparin compounds suggest serotype differences in viral entry. J Virol. 1996;70:3461–3469. doi: 10.1128/jvi.70.6.3461-3469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herold B C, Gerber S I, Polonsky T, Belval B J, Shaklee P N, Holme K. Identification of structural features of heparin required for inhibition of herpes simplex virus type 1 binding. Virology. 1995;206:1108–16. doi: 10.1006/viro.1995.1034. [DOI] [PubMed] [Google Scholar]
- 20.Herold B C, Spear P G. Neomycin inhibits glycoprotein C (gC)-dependent binding of herpes simplex virus type 1 to cells and also inhibits postbinding events in entry. Virology. 1994;203:166–171. doi: 10.1006/viro.1994.1469. [DOI] [PubMed] [Google Scholar]
- 21.Herold B C, WuDunn D, Soltys N, Spear P G. Glycoprotein C of herpes simplex virus type 1 plays a principal role in the adsorption of virus to cells and in infectivity. J Virol. 1991;65:1090–1098. doi: 10.1128/jvi.65.3.1090-1098.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hidaka Y, Okada K, Kusuhara K, Miyazaki C, Tokugawa K, Ueda K. Exanthem subitum and human herpesvirus 7 infection. Pediatr Infect Dis J. 1994;13:1010–1011. [PubMed] [Google Scholar]
- 23.Johnson D C, Ligas M W. Herpes simplex viruses lacking glycoprotein D are unable to inhibit virus penetration: quantitative evidence for virus-specific cell surface receptors. J Virol. 1988;62:4605–4612. doi: 10.1128/jvi.62.12.4605-4612.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kari B, Gehrz R. Structure, composition and heparin binding properties of a human cytomegalovirus glycoprotein complex designated gC-II. J Gen Virol. 1993;74:255–64. doi: 10.1099/0022-1317-74-2-255. [DOI] [PubMed] [Google Scholar]
- 25.Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lusso P, Secchiero P, Crowley R W, Garzino-Demo A, Berneman Z N, Gallo R C. CD4 is a critical component of the receptor for human herpesvirus 7: interference with human immunodeficiency virus. Proc Natl Acad Sci USA. 1994;91:3872–3876. doi: 10.1073/pnas.91.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Megaw A G, Rapaport D, Avidor B, Frenkel N, Davison A J. The DNA sequence of the RK strain of human herpesvirus 7. Virology. 1998;244:119–132. doi: 10.1006/viro.1998.9105. [DOI] [PubMed] [Google Scholar]
- 28.Mukai T, Hata A, Isegawa Y, Yamanishi K. Characterization of glycoprotein H and L of human herpesvirus 7. Microbiol Immunol. 1997;41:43–50. doi: 10.1111/j.1348-0421.1997.tb01171.x. [DOI] [PubMed] [Google Scholar]
- 29.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus-7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pereira L. Function of glycoprotein B homologues of the family herpesviridae. Infect Agents Dis. 1994;3:9–28. [PubMed] [Google Scholar]
- 31.Petrovskis E A, Meyer A L, Post L E. Reduced yield of infectious pseudorabies virus and herpes simplex virus from cell lines producing viral glycoprotein gp50. J Virol. 1988;62:2196–2199. doi: 10.1128/jvi.62.6.2196-2199.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfeiffer B, Berneman Z N, Neipel F, Chang C K, Tirwatnapong S, Chandran B. Identification and mapping of the gene encoding the glycoprotein complex gp82-gp105 of human herpesvirus 6 and mapping of the neutralizing epitope recognized by monoclonal antibodies. J Virol. 1993;67:4611–4620. doi: 10.1128/jvi.67.8.4611-4620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pfeiffer B, Thomson B, Chandran B. Identification and characterization of a cDNA derived from multiple splicing that encodes envelope glycoprotein gp105 of human herpesvirus 6. J Virol. 1995;69:3490–3500. doi: 10.1128/jvi.69.6.3490-3500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Portolani M, Cermelli C, Mirandola P, Di Luca D. Isolation of human herpesvirus 7 from an infant with febrile syndrome. J Med Virol. 1995;45:282–283. doi: 10.1002/jmv.1890450307. [DOI] [PubMed] [Google Scholar]
- 35.Pruksananonda P, Hall C B, Insel R A, McIntyre K, Pellett P E, Long C E, Schnabel K C, Pincus P H, Stamey F R, Dambaugh T R, Stewart J A. Primary human herpesvirus 6 infection in young children. N Engl J Med. 1992;326:1445–1450. doi: 10.1056/NEJM199205283262201. [DOI] [PubMed] [Google Scholar]
- 36.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2296. [Google Scholar]
- 37.Rostand K S, Esko J D. Microbial adherence to and invasion through proteoglycans. Infect Immun. 1997;65:1–8. doi: 10.1128/iai.65.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salahuddin S Z, Ablashi D V, Markham P D, Josephs S F, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, Gallo R C. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 39.Samuelson L C. Transgenic approaches to salivary gland research. Annu Rev Physiol. 1996;58:209–29. doi: 10.1146/annurev.ph.58.030196.001233. [DOI] [PubMed] [Google Scholar]
- 40.Secchiero P, Berneman Z N, Gallo R C, Lusso P. Biological and molecular characteristics of human herpesvirus 7: in vitro growth optimization and development of a syncytia inhibition test. Virology. 1994;202:506–512. doi: 10.1006/viro.1994.1371. [DOI] [PubMed] [Google Scholar]
- 41.Secchiero P, Berneman Z N, Sun D, Nicholas J, Reitz M S J. Identification of envelope glycoproteins H and B homologues of human herpesvirus 7. Intervirology. 1997;40:22–32. doi: 10.1159/000150517. [DOI] [PubMed] [Google Scholar]
- 42.Secchiero P, Gibellini D, Flamand L, Robuffo I, Marchisio M, Capitani S, Gallo R C, Zauli G. Human herpesvirus 7 induces the downregulation of CD4 antigen in lymphoid T cells without affecting p56Lck levels. J Immunol. 1997;159:3412–3423. [PubMed] [Google Scholar]
- 43.Secchiero P, Sun D, De Vico A L, Crowley R W, Reitz M S J, Zauli G, Lusso P, Gallo R C. Role of the extracellular domain of human herpesvirus 7 glycoprotein B in virus binding to cell surface heparan sulfate proteoglycans. J Virol. 1997;71:4571–4580. doi: 10.1128/jvi.71.6.4571-4580.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spear P G, Shieh M T, Herold B C, WuDunn D, Koshy T I. Heparan sulfate glycosaminoglycans as primary cell surface receptors for herpes simplex virus. Adv Exp Med Biol. 1992;313:341–353. doi: 10.1007/978-1-4899-2444-5_33. [DOI] [PubMed] [Google Scholar]
- 45.Tal-Singer R, Peng C, Ponce De Leon M, Abrams W R, Banfield B W, Tufaro F, Cohen G H, Eisenberg R J. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J Virol. 1995;69:4471–4483. doi: 10.1128/jvi.69.7.4471-4483.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka K, Kondo T, Torigoe S, Okada S, Mukai T, Yamanishi K. Human herpesvirus 7: another causal agent for roseola (exanthem subitum) J Pediatr. 1994;125:1–5. doi: 10.1016/s0022-3476(94)70113-x. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka-Taya K, Kondo T, Mukai T, Miyoshi H, Yamamoto Y, Okada S, Yamanishi K. Seroepidemiological study of human herpesvirus-6 and -7 in children of different ages and detection of these two viruses in throat swabs by polymerase chain reaction. J Med Virol. 1996;48:88–94. doi: 10.1002/(SICI)1096-9071(199601)48:1<88::AID-JMV14>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 48.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 49.Whitbeck J C, Peng C, Lou H, Xu R, Willis S H, de Leon M P, Peng T, Nicola A V, Montgomery R I, Warner M S, Soulika A M, Spruce L A, Moore W T, Lambris J D, Spear P G, Cohen G H, Eisenberg R J. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J Virol. 1997;71:6083–6093. doi: 10.1128/jvi.71.8.6083-6093.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyatt L S, Frenkel N. Human herpesvirus 7 is a constitutive inhabitant of adult human saliva. J Virol. 1992;66:3206–3209. doi: 10.1128/jvi.66.5.3206-3209.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wyatt L S, Rodriguez W J, Balachandran N, Frenkel N. Human herpesvirus 7: antigenic properties and prevalence in children and adults. J Virol. 1991;65:6260–6265. doi: 10.1128/jvi.65.11.6260-6265.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yadav M, Nambiar S, Khoo S P, Yaacob H B. Detection of human herpesvirus 7 in salivary glands. Arch Oral Biol. 1997;42:559–567. doi: 10.1016/s0003-9969(97)00049-6. [DOI] [PubMed] [Google Scholar]
- 53.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;i:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]
- 54.Yoshikawa T, Asano Y, Kobayashi I, Nakashima T, Yazaki T, Suga S, Ozaki T, Wyatt L S, Frenkel N. Seroepidemiology of human herpesvirus 7 in healthy children and adults in Japan. J Med Virol. 1993;41:319–323. doi: 10.1002/jmv.1890410412. [DOI] [PubMed] [Google Scholar]