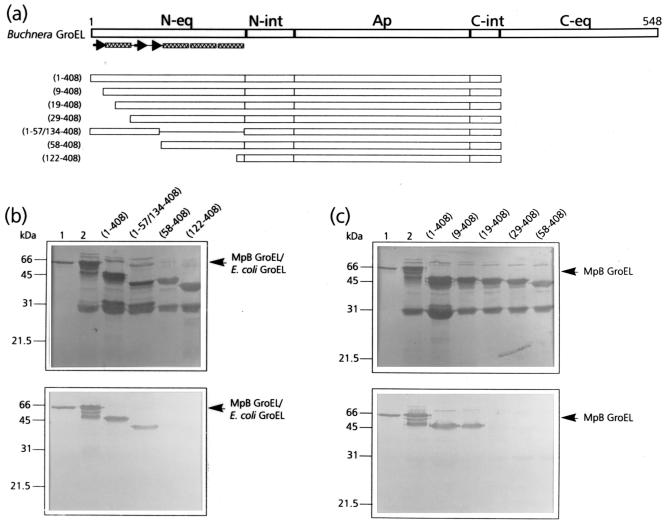
FIG. 1.
Mapping of the N-terminal PLRV-binding site by virus overlay assays of deletion derivatives of MpB GroEL. (a) Schematic representation of MpB GroEL deletion mutants. The numbers in parentheses correspond to the positions of amino acid residues of MpB GroEL (17) and mark the borders of the deletion mutants. N-eq, N-terminal region of the equatorial domain; N-int, N-terminal region of the intermediate domain; Ap, apical domain; C-int, C-terminal region of the intermediate domain; C-eq, C-terminal region of the equatorial domain. Secondary structural elements are indicated by boxed sine waves (α-helices) and arrows (β-strands). (b) Virus overlay assay showing that the first 57 amino acid residues are involved in PLRV binding (bottom) and amido black-stained blot (top). (c) Virus overlay assay showing that residues 10 to 18 are involved in virus binding (bottom) and amido black-stained blot (top). Lanes 1, wild-type MpB GroEL isolated from M. persicae; lanes 2, recombinant MpB GroEL. All other lanes contain the indicated deletion mutants of MpB GroEL as depicted in panel a. The positions of MpB GroEL and E. coli GroEL are indicated.