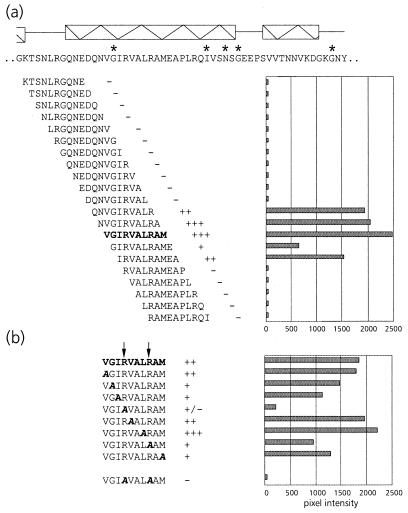
FIG. 5.
(a) Schematic representation of the virus overlay assays of decameric peptides corresponding to amino acid residues 423 to 476 of the C-terminal region of the equatorial domain of MpB GroEL. The results of the first 21 peptides are shown; no PLRV binding to any of the other peptides in this region was detected. Secondary structural elements are indicated by boxed sine waves (α-helices). Conserved sequences in GroEL/Hsp60 sequences are indicated by asterisks. (b) Alanine scanning of the decameric peptide with the strongest binding capacity as indicated in panel a (in boldface). The affinity of PLRV for the peptides has been quantified using Molecular Analyst software (histograms) and is interpreted as follows: +++, high affinity; ++, intermediate affinity; +, low affinity; −, no PLRV binding detected. Arrows indicate residues critical for binding PLRV.