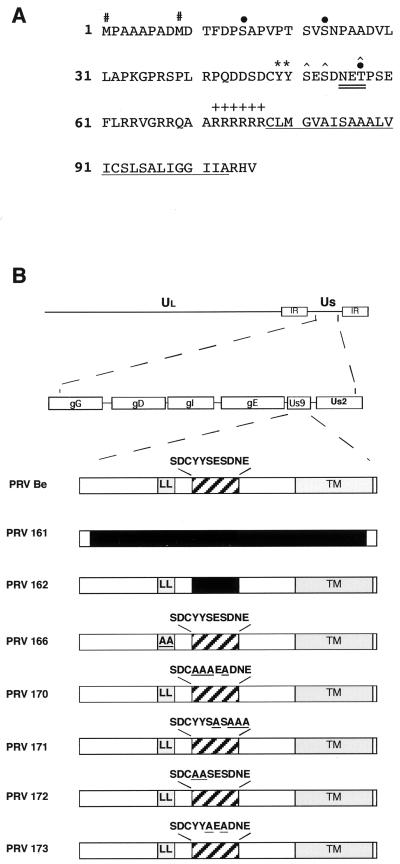
FIG. 1.
(A) Amino acid sequence of the Us9 open reading frame product. The two in-frame methionine residues (#) and the potential tyrosine kinase phosphorylation sites (∗) are indicated. The consensus casein kinase I sites [(S/T)X2–3(S/T)X] (31) each are each indicated by a solid circle above the S or T boldfaced here, and the consensus casein kinase II sites [X(S/T)XX(D/E)] are each marked with a caret above the S or T (PROSITE pattern). The potential N-linked glycosylation [NX(S/T)] sequence is double underlined. The putative transmembrane domain is underlined, and the surrounding basic residues are indicated by plus signs. (B) PRV genome and maps of viruses used in this study. The PRV genome is shown on the first line, and the unique short region (Us) is expanded on the second line. The Us9 proteins from the various viruses used in this study are diagrammed below. The dileucine endocytosis motif (LL) and the transmembrane domain (TM) are indicated by shaded boxes. The acidic domain is indicated by a hatched box with the amino acid sequence given above. All amino acid substitutions are underlined, and deletions are indicated by solid boxes. PRV Be is the wild-type strain used in this study. PRV 161 contains a 258-bp deletion in the Us9 open reading frame. PRV 162 contains a 10-amino-acid deletion removing a conserved acid motif in the Us9 cytoplasmic tail. PRV 166 contains a leucine-to-alanine substitution at amino acids 30 and 31 in the Us9 protein. PRV 170 (Y49–50A S51A S53A), PRV 171 (E52A D54A N55A E56A), PRV 172 (Y49–50A), and PRV 173 (S51A S53A) contain alanine substitutions in the conserved acidic domain in the Us9 cytoplasmic tail (see the text for details).