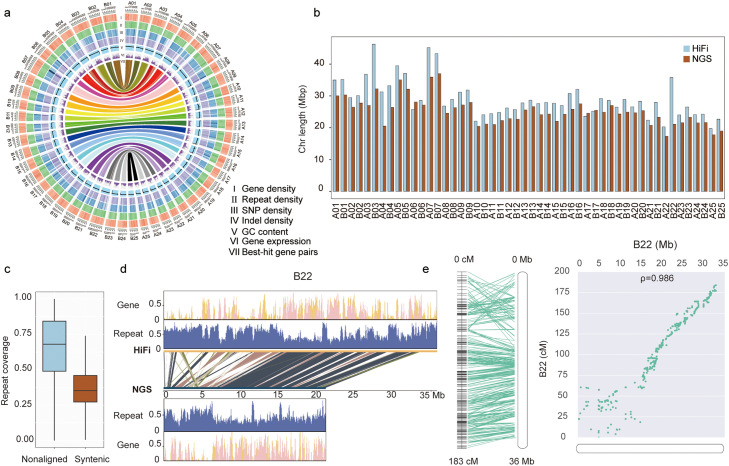
Fig. 1.
Characterization of the common carp HiFi assembly. (a) Features of homoeologous chromosomes in the HiFi genome, CC 3.0. From outer to inner: I, gene density; II, repeat density; III, SNP density; IV, indel density; V, GC content; VI, gene expression based on adult muscle tissue; VII, best-hit gene pairs between two subgenomes. Gene density and repeat distribution were calculated within each 100-kb sliding window. SNP and indel density were based on previously published resequencing data for Yellow River carp [7]. (b) Chromosome length comparison between the HiFi and NGS versions of common carp genomes. (c) Repeat coverage of syntenic and nonaligned regions between the HiFi and NGS version genomes. (d) Alignment of chromosome B22 between the HiFi and NGS genomes. The center shows syntenic blocks, with orange lines for gene density, and purple for repeat density. (e) Example showing the relationship between the HiFi genome and linkage map [68]. The linkage map is on the left.