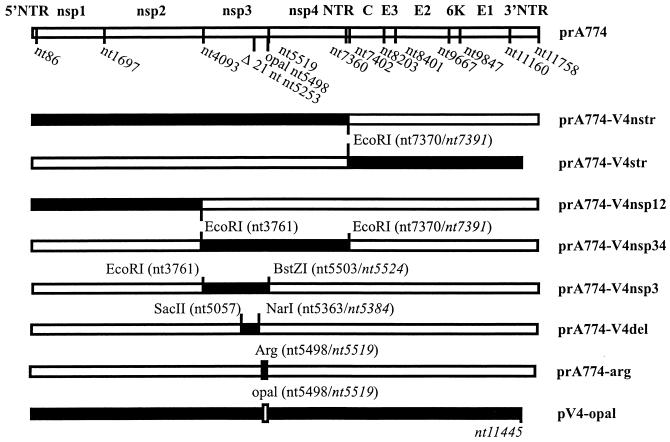
FIG. 1.
Schematic illustration of the full-length SFV cDNA constructs used as a source for infectious RNA. Solid bars indicate SFV4 cDNA, and open bars represent the A7(74) genome. The pGEM1-derived vector (24) is not shown. The 21-nt deletion in prA774 alters the numbering of nucleotides with regard to SFV4. SFV4-specific nucleotide position numbers are given in italics. The 3′ NTR of SFV4 is 334 nt shorter than that of rA774 (34). prA774 is the cDNA clone of A7(74). prA774-V4nstr was obtained by combining the 5′NTR and nonstructural genes of SFV4 with the structural genes and 3′NTR of rA774. prA774-V4str is reciprocal to prA774-V4nstr. In prA774-V4nsp12, a fragment containing the 5′ NTR, nsp1, and nsp2 of SFV4 was introduced into prA774, and similarly, in prA774-V4nsp34 and prA774-V4nsp3, the indicated genes were used to replace the corresponding regions in rA774. In prA774-V4del, a short fragment of SFV4 nsp3 containing the 21 nt deleted in rA774 has been inserted into the corresponding region of rA774. prA774-arg differs from rA774 only by having an arginine codon at the opal position. pV4-opal is identical to SFV4 except that the arginine has been mutated to opal.