FIGURE 1.
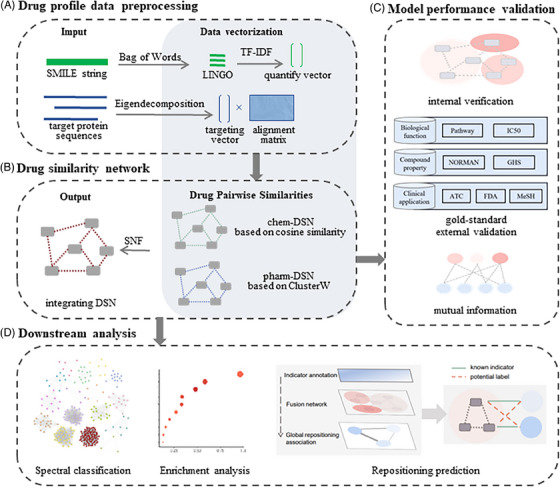
The overall flowchart of the proposed framework. (A) In the first part, we preprocess structural data and target protein sequences based on 276 anti‐tumor drugs containing target protein, PubChem CID and pathway annotations in GDSC to construct eigenvectors or eigen decomposition matrices that can be used for similarity network analysis. (B) In the second part, we constructed two structure‐based single‐property DSNs by vector space model and multiple sequence alignment denoted as chem‐DSN and pharm‐DSN, respectively, and fused them into multi‐scale network, named iDSN. (C) In the third part, we used the drug classification notes on DrugBank and PubChem as the gold standard to test the classification effect of DSN according to the adjusted Rand index. (D) In the fourth part, 276 drugs included in the iDSN were grouped into 16 clusters (Cluster 1–Cluster 16) and performed a systematic statistic analysis for annotating iDSN clusters from various perspectives to guide drug repositioning. Possible drug repositioning applications were obtained based on high‐similarity drug pairs or the occurrence of unexpected drugs within iDSN clusters.
