Figure 1. Overview of the cohorts and analysis pipeline (a) and genetic correlations among the sites (b).
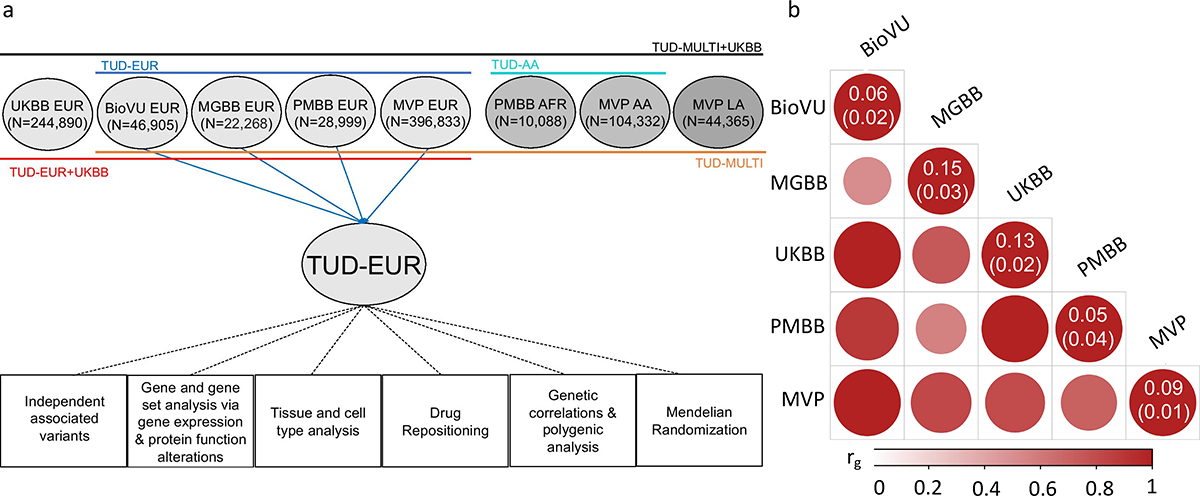
(a) We conducted independent GWAS of TUD cases and controls in individuals of European (EUR) ancestry across four PsycheMERGE sites (BioVU, MGBB, PMBB, and MVP) and performed a GWAS meta-analysis (“TUD-EUR”); these summary results were used for all secondary analyses. For African American (AA), we conducted GWAS meta-analysis of TUD cases and controls from the PMBB and MVP cohorts (“TUD-AA”). For Latin American (LA), we conducted GWAS of TUD cases and controls from the MVP cohort. Next, we performed a multi-ancestral GWAS meta-analysis (“TUD-multi”), which combined the results from all seven cohorts. We also obtained summary statistics from UKBB, which used a less stringent case definition in individuals of EUR ancestry and performed a GWAS meta-analysis within EUR individuals (“TUD-EUR+UKBB”) and across ancestries (“TUD-multi+UKBB”). Supplementary Table 2 summarizes the datasets used for the analyses. We subjected the TUD-EUR summary statistics to several secondary analyses to characterize the genetic architecture of TUD. (b) LDSC genetic correlations (rg) for TUD between EUR sites were positive and high, ranging from 0.51 to unity (two-sided p-values are provided in Supplementary Table 6), with most confidence intervals overlapping (Supplementary Figure 1). LDSC genetic correlation for TUD between the two AA samples was strongly positive (rg=0.93) but not significant (p=0.45). LDSC SNP-heritability estimates (h2SNP 5–15%) are shown in the diagonal. UKBB=UK Biobank, BioVU=Vanderbilt University Medical Center’s biobank, MGBB=Mass General Brigham Biobank, PMBB=Penn Medicine Biobank, MVP=Million Veteran Program.
