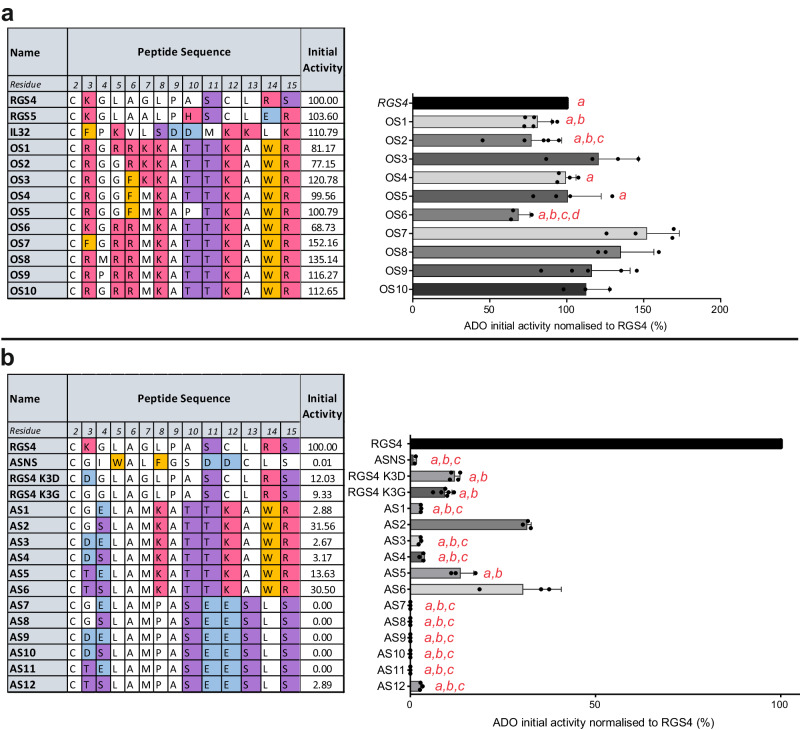
Fig. 3. In vitro testing of proposed ADO optimum substrate (OS) and anti-substrate (AS) peptides.
a Sequences of ADO optimum substrate (OS) peptides and initial activity of HsADO with OS peptides. Significant differences (p < 0.05) from 1-way ANOVA results summarised on graph: a = lower than OS7; b = lower than OS8; c = lower than OS3; d = lower than OS9 and OS10. Full 1-way ANOVA results and p-value ranges available in Supplementary Data 3. b Sequences of ADO anti-substrate (AS) peptides and initial activity of HsADO with AS peptides and RGS4 K3D/G. ASNS was included as a negative control for ADO activity. Significant differences (p < 0.05) from 1-way ANOVA results summarised on graph: a = lower than AS2; b = lower than AS6; c = lower than AS5; ASNS, RGS4 K3D/G and AS1-12 all lower than RGS4. Full 1-way ANOVA results available and p-value ranges in Supplementary Data 4. For (a, b), initial activity is turnover of peptide after 1-min incubation with HsADO normalised to that of RGS42-15 positive control peptide (%). Data represent mean ± standard deviation; n ≥ 3. Amino acids are colour coded according to chemical properties: basic (H/K/R) = red; acidic (D/E) = blue; aromatic (F/W) = orange; polar (S/T) = purple. Source data are provided as a Source Data file.