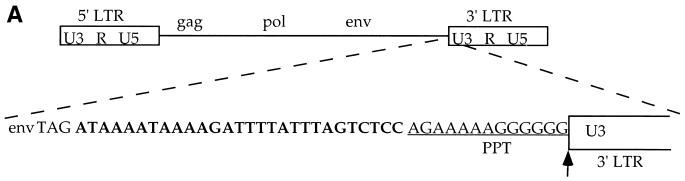
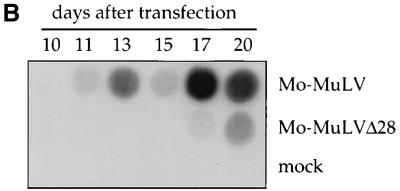
FIG. 1.
Structure and replication of Mo-MuLV and Mo-MuLVΔ28. (A) Schematic representation of the organization of the Mo-MuLV genomic DNA (upper drawing), and an expanded view of the PPT with the upstream untranslated sequence (lower drawing). The U3, R, and U5 regions in the LTRs are shown (open boxes). The PPT sequence is underlined, and the sequence of the 28 bases deleted in Mo-MuLVΔ28 is shown in bold letters. This sequence is located 3′ to the envelope stop codon (envTAG) and 5′ to the PPT sequence. The vertical arrow indicates the RNase H cleavage site, necessary for the formation of the correct 5′ LTR edge during reverse transcription. (B) Viral spread in Rat2-2 cells. Rat 2-2 cells were transiently transfected with DNA expressing the indicated viruses, and the cultures were passaged at 1:20 dilution every 3 to 4 days. The decrease seen at day 15 is due to a transfer the previous day. An exogenous RT assay was performed on supernatants collected on the indicated days after transfection. A control transfection was performed without adding plasmid DNA (mock).