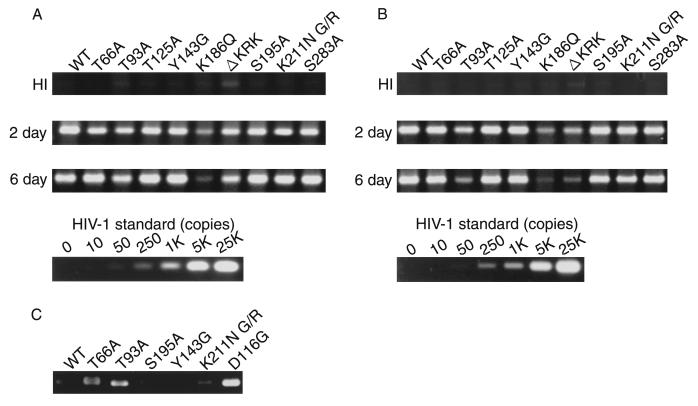
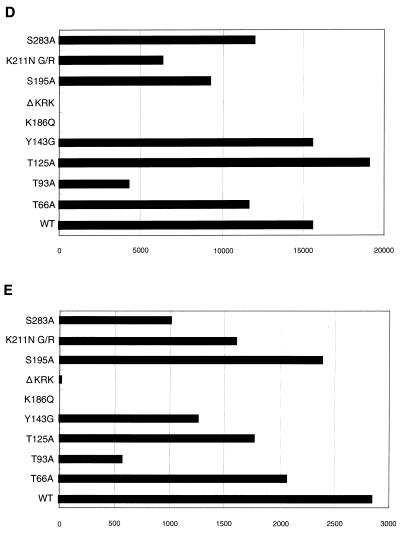
FIG. 3.
Analysis of IN mutants in dividing cells. Each virus was prepared by cotransfection of COS cells with pNL43lucΔenv vector and pJD-1. The DNase-treated supernatants containing ∼10 ng of p24 were inoculated into RD cells or PBLs. At 2 or 6 days postinfection, as indicated on the left, the entire cell culture was harvested. Total DNA was extracted from infected RD cells and subjected to PCR analysis with the primer pairs for R/U5 (A) and R/gag (B) and the two-LTR circle (C). For HIV-1 DNA standards, 50 to 100,000 copies of linearized HIV-1 JR-CSF DNA were amplified in parallel. Amplified products were resolved on 2% agar gel and visualized by Syber-Green staining (FMC Bioproduct). Virus treated at 65°C for 30 min prior to inoculation was used as a heat-inactivated control (HI). After 4 days of infection, the entire cells were harvested and washed with PBS. The cell pellets were resuspended with 200 μl (for RD cells) (D) and 100 μl (for PBLs) (E) of luciferase lysis buffer (Promega Corp.). Ten microliters of each cell lysate was subjected to luciferase assay as described in Materials and Methods. Luciferase activities are shown in units per microliter.