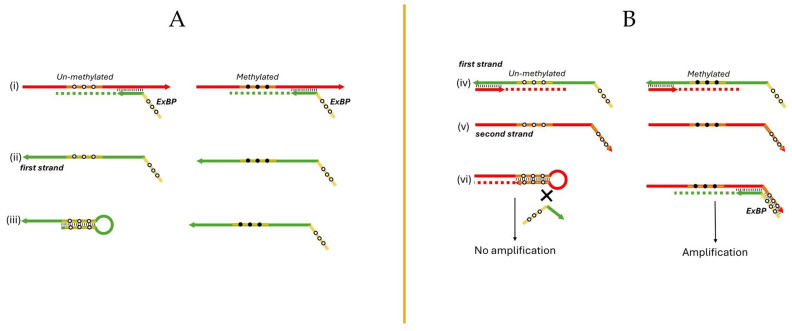
Figure 1.
Schematic of the semi-nested real-time PCR assay with the ExBPs. (A) features extendable blocking probes (ExBPs) at the 5′ ends of forward primers (Panel (i)), which are designed to anneal to target regions free of CpG sites, allowing binding to both methylated and unmethylated sequences. Panel (ii) illustrates the synthesis of the ‘first strand’, initiated from the primer containing the ExBPs, with Panel (iii) depicting the specific hybridization of ExBPs to unmethylated CpG sites, and its inability to align with methylated sequences. (B) demonstrates the extension process of the PCR primer forming a complementary “second strand” from the first strand (Panels (iv) and (v)). Panel (vi) highlights the formation of a stable hairpin structure at the 3′ end of this second strand if derived from unmethylated DNA, effectively preventing it from acting as a template in subsequent PCR cycles/stages, and thereby enhancing the selective amplification of methylated DNA sequences.