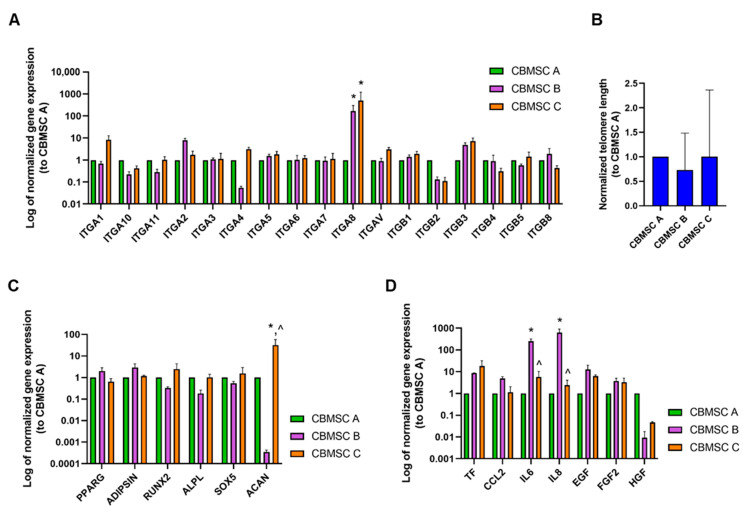
Figure 3.
For information only characterization of cord blood-derived mesenchymal stromal cells. (A) Histograms show gene expression of integrins normalized to CBMSC A; data are presented as mean ± standard deviation (SD) (n = 3). Statistical analysis was by two-way ANOVA followed by Tukey’s multiple comparisons post-hoc test; * p < 0.05 vs. CBMSC A. (B) Histograms show telomere length normalized to CBMSC A; data are presented as mean ± SD (n = 3). Statistical analysis was by non-parametric Kruskal-Wallis test followed by Dunn’s multiple comparisons post-hoc test. (C) Histograms show transcript levels of differentiation-associated genes normalized to CBMSC A; data are presented as mean ± SD (n = 3). Statistical analysis was by two-way ANOVA followed by Tukey’s multiple comparisons post-hoc test; * p <0.05 vs. CBMSC A, ^ p < 0.05 vs. CBMSC B. (D) Histograms show gene expression of secreted proteins normalized to CBMSC A; data are presented as mean ± SD (n = 3). Statistical analysis was by two-way ANOVA followed by Tukey’s multiple comparisons post-hoc test; * p < 0.05 vs. CBMSC A, ^ p < 0.05 vs. CBMSC B. Abbreviation list: CBMSC, cord blood mesenchymal stromal cells; SD, standard abbreviation.