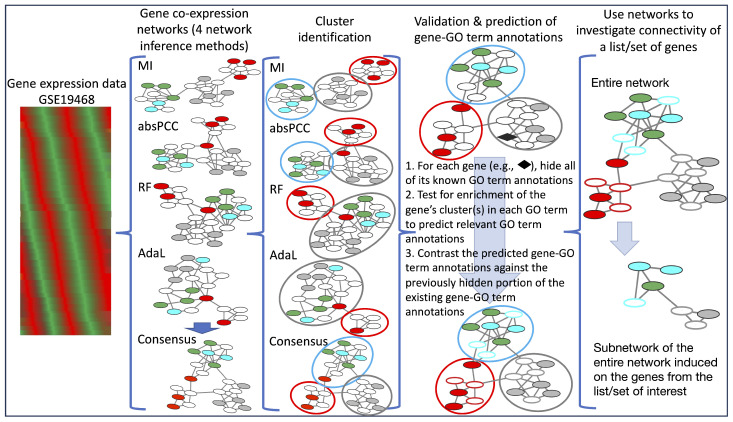
Figure 1.
Summary of our generalizable framework for gene co-expression network construction, validation, and biological application. We use four network inference methods (MI, absPCC, RF, and AdaL) to construct gene co-expression networks of P. falciparum; note that in the illustration, nodes having the same color indicates that they are annotated by the same GO term. We also generate a Consensus network by combining the four individual networks. We use two clustering methods to identify gene clusters’ (hypothesized functional modules) in each network. Then, we use crossvalidation to examine how well each network’s clusters correspond to existing GO terms. In other words, GO term annotations are predicted for all genes whose clusters are statistically significantly enriched in at least one GO term, and the predicted gene–GO term annotations are then contrasted against a previously hidden portion of the existing ones. The Consensus network is used to investigate the connectivity of a gene list/set; in our biological application, we investigate lists of genes that interact with three proteins involved with the biological process of endocytosis.