Figure 2.
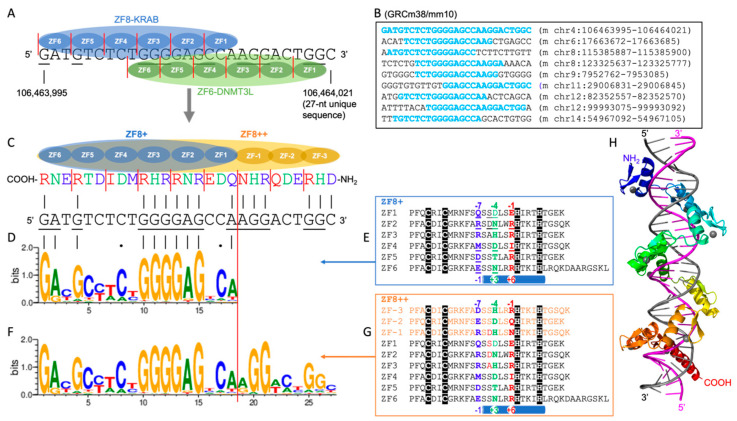
Improved specificity based on ZF8. (A) Overlap between ZF8-KRAB and ZF6-DNMT3L. The 18 bps recognized by ZF8-KRAB and the 18 bps recognized by ZF6-DNMT3L overlap by 10 bps. Together, they recognize a 27 bp segment. (B) Several shorter sequences, under 27 nt, display partial matches on other chromosomes of the mouse genome (GRCm38/mm10). (C) The design of an expanded nine-finger protein. The protein sequence from the NH2 to COOH termini (right-to-left) runs antiparallel to that of the DNA sequence from the 5′ to 3′ ends (left-to-right). (D,E) Improved specificity of ZF8+ fusion protein (sequence logo in (D)) and the corresponding protein sequence with altered residues underlined (E). (F,G) Improved specificity of ZF8++ fusion protein (sequence logo in (F)) and the corresponding protein sequence of the nine-finger array (G). Note that the sequence-based numbering (−1, −4, and −7) and the structure-based numbering (+6, +3, and −1) are provided above and below the sequences, respectively. (H) An AlphaFold3 prediction of ZF8++ in a complex with DNA with the nine ZF units (colored from blue to red), and the DNA recognition strand (magenta).