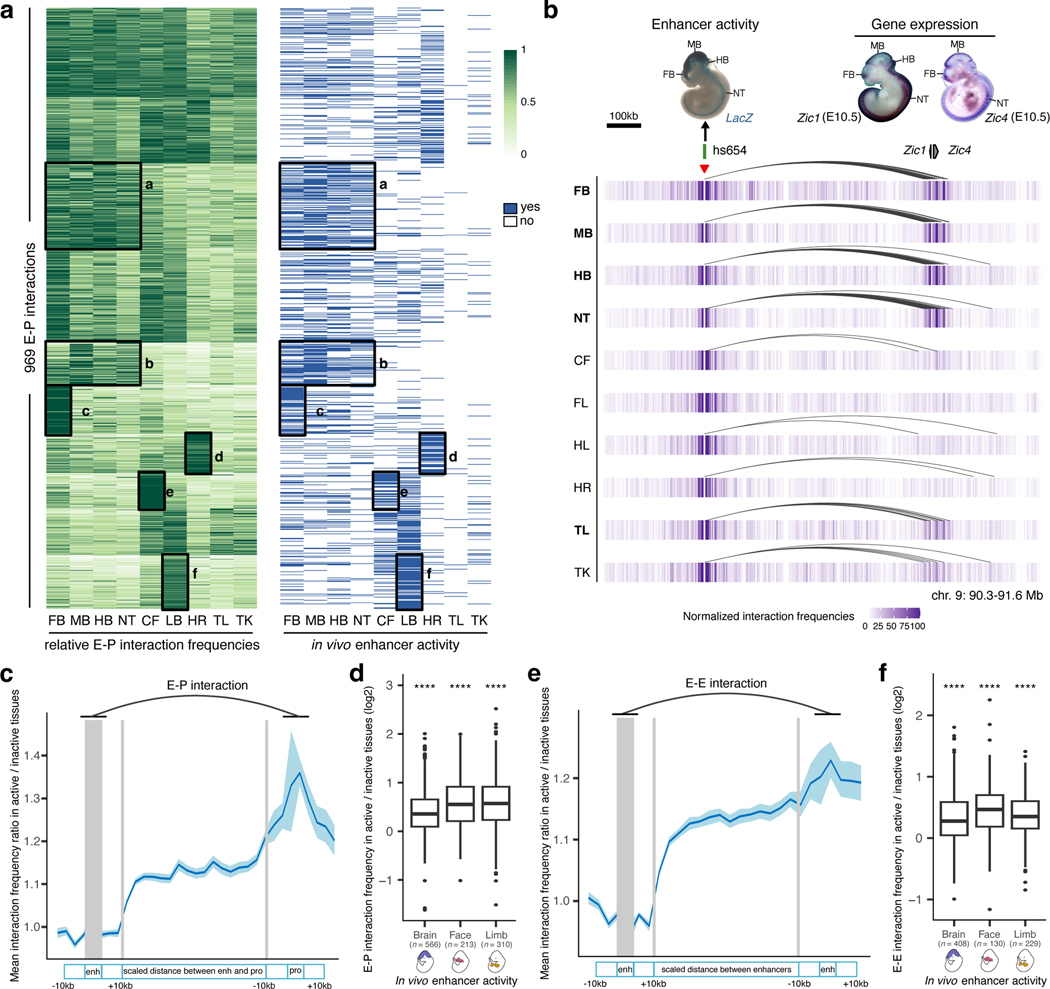
Fig. 4: Tissue specificity of developmental enhancer interactions.
a, Heatmap showing relative E–P chromatin interaction frequencies (scaled to the max value among tissues in each E–P interaction, green) and the in vivo enhancer activities (blue) of 969 E–P chromatin interactions. k-means clustering (k = 10) was performed on interaction frequencies. The six highlighted tissue-specific interaction clusters match in vivo enhancer activities. b, Interaction profiles across 10 tissues centered on the hs654 enhancer (red arrowhead indicates capture Hi-C viewpoint). The top left shows hs654 enhancer activity in a transgenic mid-gestation (E11.5) mouse embryo. Top right images show Zic1 and Zic4 mRNA whole-mount in situ hybridization (WISH) at E10.5 (Images reproduced with permission from Gene Expression Database (GXD; Zic4)35 and Embrys database (http://embrys.jp; Zic1). Heatmaps with normalized interaction frequencies in each of the 10 tissues are shown below. Curved lines indicate significant interactions. c, e, Average ratio of E–P or E–E interaction frequency between active and inactive tissues based on the analysis of 946 E–P or 640 E–E chromatin interactions are shown (see Methods for details of normalization procedure). Light blue shading indicates 95% confidence intervals estimated by non-parametric bootstrapping. d, f, Average ratio of E–P or E–E interaction frequency between active and inactive tissues for enhancers active in brain, face and limb (see Extended Data Fig. 6a and Extended Data Fig. 8b for other tissues). The P values for E–P interactions are 5.07×10−61 (Brain), 6.1×10−28 (Face), 6.21×10−43 (Limb). The P values for E–E interactions are 3.3×10−38 (Brain), 1×10−17 (Face), 1.5×10−29 (Limb). FB, forebrain. MB, midbrain. HB, hindbrain. CF, craniofacial mesenchyme. HR, heart. FL, forelimb. HL, hindlimb. TK, trunk. TL, tail. NT, neural tube. For the boxplots in panels d and f, the central horizontal lines are the median, with the boxes extending from the 25th to the 75th percentiles. The whiskers further extend by ±1.5 times the interquartile range from the limits of each box.