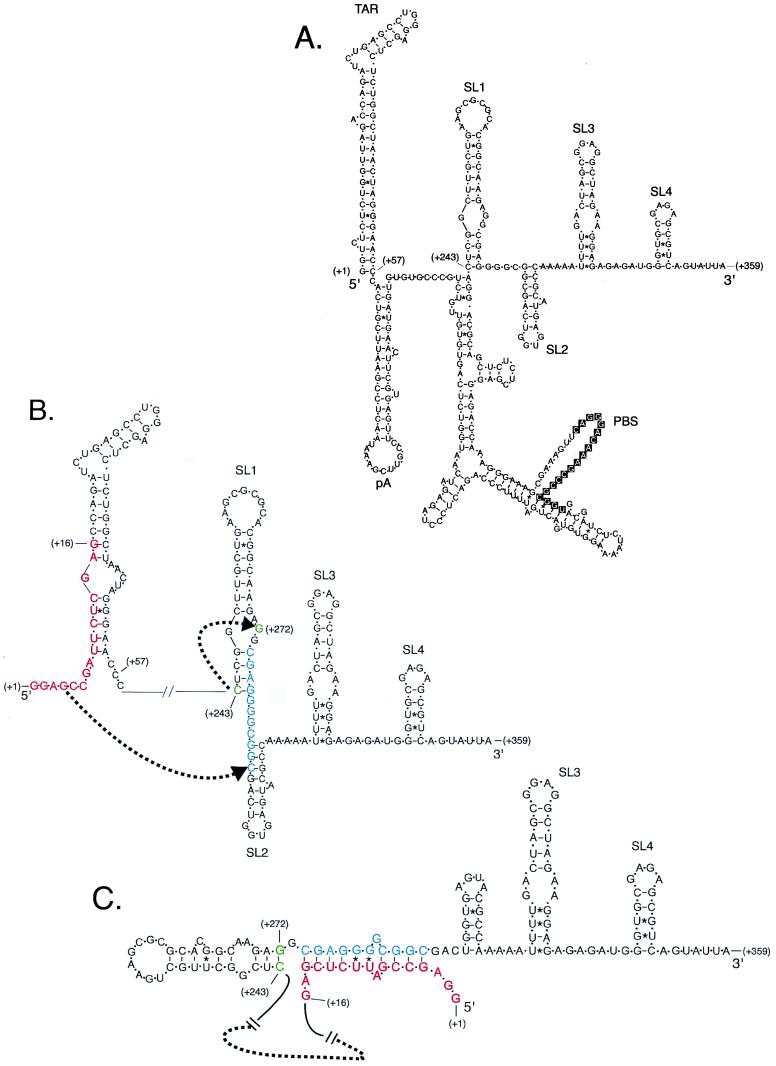
FIG. 7.
Computer model of potential HIV-1 RNA secondary structures in the presences of stem-destabilizing clustered mutations. (A) HIV-1 RNA secondary structure predicted from the wild-type HIV-1 HXB2 sequence from position +1 to +359 using M-fold version 3.0. Stem-loop structures corresponding to TAR, pA, SL1, SL2, SL3, and SL4 as well as the primer binding site (PBS) are indicated. (B) Predicted secondary structure of TAR RNA (positions +1 to +57) showing the clustered mutation Xho+10 (10) indicated by red letters. HIV-1 nt +274 to +284 (shown in blue) can base pair to sequences in Xho+10. A dotted arrow indicates a shift by cytidine +243 (green) to base pair with guanine +272 (green). (C) The new secondary structure maintains TAR and pA stem-loop structures (not shown), SL3, and SL4 but eliminates SL1 and SL2. The free energy of this structure is −40.2 kcal/mol compared to the structure shown in panel B, from position +243 to +359, with free energy of −33.9 kcal/mol calculated by M-fold version 3.0 using standardized parameters (36).