Fig. 2. Validation of TF predictions.
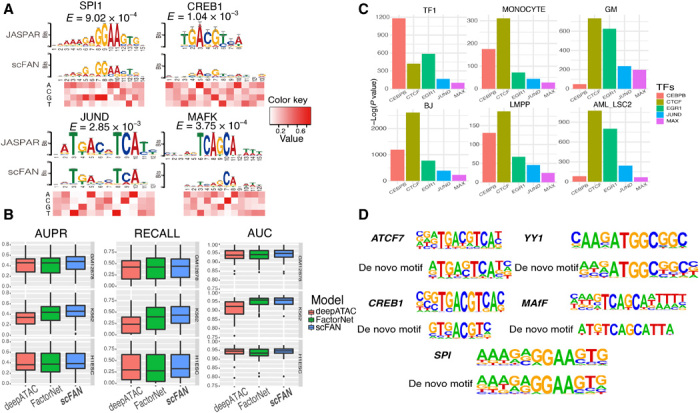
scFAN can predict both bulk and single-cell TF binding. (A) Four convolutional kernels that matched with four known motifs derived from JASPAR database. The heatmap denotes the value of each nucleotide corresponding to the above position. (B) Box plot of the performance of the pretrained model and two other models predicting bulk cell TF binding on the same dataset. (C) Enrichment analysis of the five predicted most active TFs from six randomly chosen cells. scFAN predicts the most likely TF per bin and adds up the number of times each TF is the highest predicted TF. Homer takes all the candidate peaks that need to be predicted and generates the enrichment analysis. All these TFs were significantly enriched in all these peaks. (D) Several example regions were used for enrichment analysis. scFAN was used to predict these regions’ most active TFs, which are ATCF7, YY1, CREB1, MAFF, and SPI. De novo matched motifs were compared to known motifs from Homer.
