Fig. 6. Loss of Dcaf1 impairs ribosome biogenesis and activates RP-Mdm2-p53 checkpoint pathway.
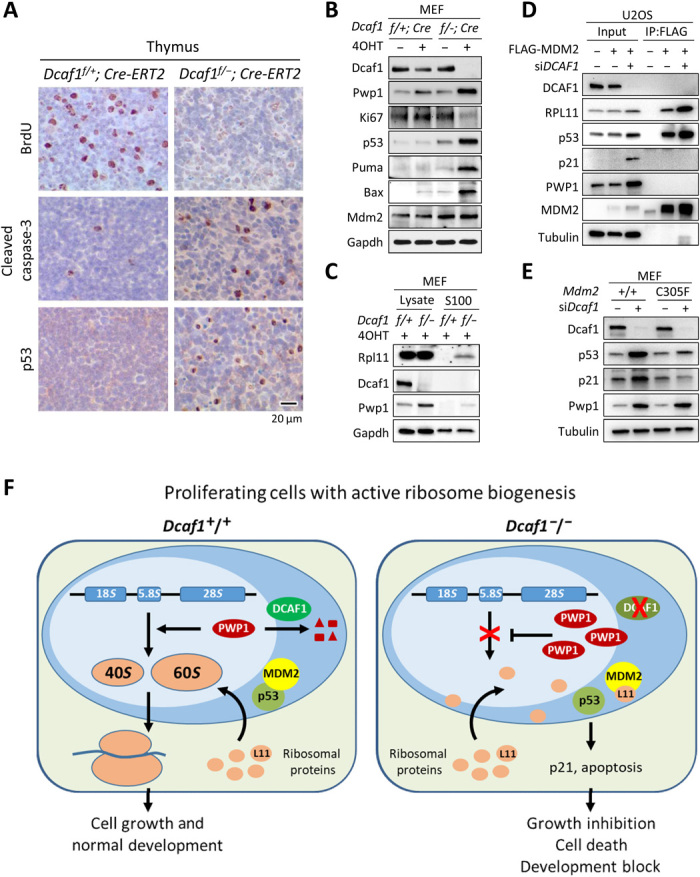
(A) IHC staining of BrdU, cleaved caspase-3, and p53 from sections of formalin-fixed, paraffin-embedded thymus tissue following tamoxifen treatment. Positive staining is indicated by brown color; sections were counterstained with hematoxylin. (B) Dcaf1f/+;Cre-ERT2 and Dcaf1f/−;Cre-ERT2 MEFs were treated with 200 nM 4OHT for 3 days. (C) 4OHT-treated Dcaf1f/+;Cre-ERT2 and Dcaf1f/−;Cre-ERT2 MEFs were homogenized in hypotonic buffer (designated as total cell lysates). Ribosomes were pelleted from lysates by ultracentrifugation at 100,000g for 3 hours at 4°C, and the ribosome-free supernatants were collected (designated as the S100 fractions). (D) U2OS cells stably expressing FLAG-tagged MDM2 or empty vector were transfected with siRNAs targeting DCAF1 or control siRNAs. The interactions between MDM2 and RPL11 proteins were determined by co-IP analysis. (E) Activation of p53 by loss of DCAF1 depends on intact RP-MDM2-p53 pathway. Primary Mdm2+/+ or Mdm2C305F/C305F MEFs were transfected with siRNAs targeting Dcaf1 or control siRNAs. (F) Schematic model for the function of DCAF1-based E3 ubiquitin ligase in the regulation of PWP1 protein, ribosome biogenesis, cell proliferation, and development.
