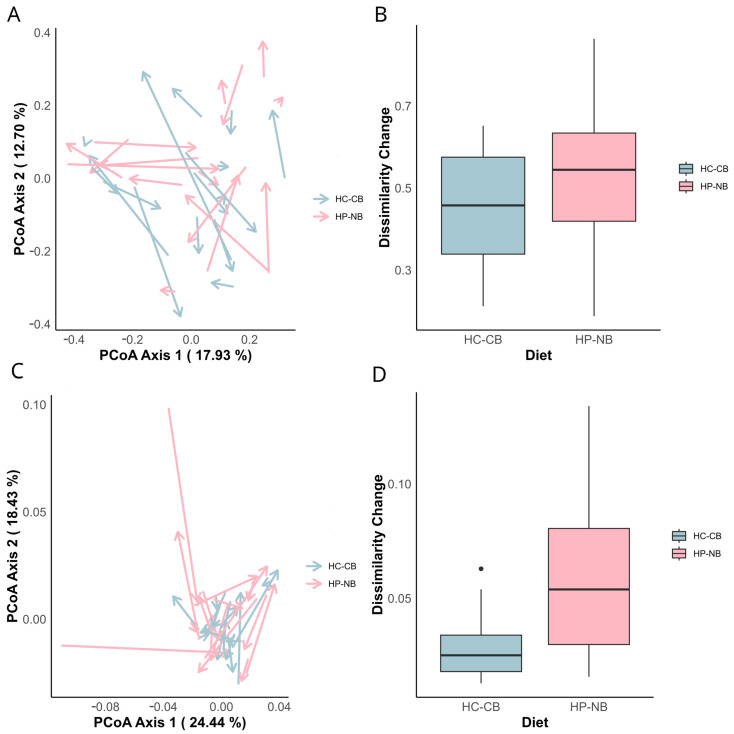
Figure 3.
Shift in bacterial community composition and KEGG level 2 metabolic pathways (Bray–Curtis dissimilarity) for the shotgun metagenome dataset (restricted to participants with overweight and prediabetes, n = 32). (A,C) Principal coordinate analysis (PCoA) plots depict changes in the bacterial community (genus level) (A) and KEGG level 2 metabolic pathways (C) between CID 1 baseline and CID 5 end of intervention. Samples are colour coded by diet group (HP-NB: red; HC-CB: blue). Arrows represent the direction and magnitude of change in the bacterial community from CID 1 to CID 5. PERMANOVA analysis revealed no statistical significance by CID at the genus level (HP-NB p = 0.89; HC-CB p = 0.44) or at KEGG level 2 (HP-NB p = 0.31; HC-CB p = 0.998). (B,D) Box and whisker plots showing the dissimilarity change (Bray–Curtis dissimilarity) between CID 1 and CID 5 for each diet group: (B) dissimilarity change in the bacterial community (genus level); (D) dissimilarity change in KEGG level 2 metabolic pathways. Outliers are represented as individual dots.