Abstract
Breast cancer is the most common malignancy in the female sex; although recent therapies have significantly changed the natural history of this cancer, it remains a significant challenge. In the past decade, evidence has been put forward that some oncogenic viruses may play a role in the development of sporadic breast cancer; however, data are scattered and mostly reported as sparse case series or small case–control studies. In this review, we organize and report current evidence regarding the role of high-risk human papillomavirus, mouse mammary tumor virus, Epstein–Barr virus, cytomegalovirus, bovine leukemia virus, human polyomavirus 2, and Merkel cell polyomavirus in the pathogenesis of breast cancer.
Keywords: breast cancer, carcinogenesis, viral carcinogenesis, HPV, MMTV, EBV, CMV, JCV, MCPYV
1. Introduction
Since the first discovery of Rous sarcoma virus (RSV) and mouse mammary tumor virus (MMTV) as causative agents of cancer in animals in 1911 [1] and 1936 [2], the role of infectious agents as etiological agents of cancer has been studied and confirmed for several entities. In 2018, an analysis of the GLOBOCAN database reported that 13% of cancer was associated with infections, with Helicobacter pylori, human papillomavirus (HPV), hepatitis B virus (HBV) and hepatitis C (HCV) virus reported as the primary causative agents [3].
Viruses are recognized to cause or contribute to several human cancers; the list includes hepatocellular carcinoma (HBV and HCV), cervical squamous cell carcinoma and adenocarcinoma, head and neck squamous cell carcinoma (HPV), Burkitt’s lymphoma, nasopharyngeal carcinoma and a subset of gastric cancer [Epstein–Barr virus (EBV)], adult T-cell leukemia [human T-cell leukemia virus (HTLV1)], Kaposi’s sarcoma [human herpesvirus 8 (HHV8)], and Merkel cell carcinoma [Merkel cell polyomavirus (MCPyV)].
Viruses can participate in oncogenesis by different means, such as direct transcription of oncogenic proteins, by contributing to a chronic inflammatory milieu, and by causing genomic alterations and instability in the infected cells.
With cancer recognized as a multifactorial disease, any external factor that can be identified and acted upon, as primary or secondary prevention, becomes important to reduce cancer risk.
Since their introduction, Bradford Hill’s criteria [4] have been used to assess causality in epidemiology; there are nine items in those criteria: strength of association, consistency of association, specificity, temporality, biological gradient (dose–response relationship), plausibility, coherence, experiment, and analogy.
Therefore, in order to establish if a virus can be considered a causative agent of cancer, epidemiological studies need to be performed on a large scale, to identify the virus in the cancer cells, and consistently reproduce the results across regions and populations.
Breast cancer is the most common cancer in women; in 2022, there were 2.3 million of women diagnosed with breast cancer [5], and a few risk factors are known: female sex, genetic predisposition, increasing age, obesity, alcohol and tobacco use, family history, radiation exposure, reproductive history, early menarche and late first pregnancy, and postmenopausal hormone therapy. However, half of breast cancer cases occur in women with no known risk factor, except from age and female sex, and it is therefore likely that some risk factors are still undiscovered.
In this context, there is much interest in the scientific community in identifying new risk factors, in the hope of significantly impacting such a common cancer.
The possibility that viruses may have a role in the development of breast cancer has been investigated for some years for several viral agents, and many conflicting results have been reported in the literature.
In this review, we collect and examine the hypothesized pathogenetic mechanisms of viral breast carcinogenesis regarding HPV, MMTV, EBV, human cytomegalovirus (CMV), bovine leukemia virus (BLV), human polyomavirus 2 (JCV), and Merkel cell polyomavirus (MCPyV), we report the prevalence of these virus in breast cancer specimens, and we systematically review the results of studies that also included controls in their design to provide an up-to-date account of the progress that has been made in this field of study in recent years.
2. Materials and Methods
We performed a systematic review of the studies including controls in their study design present in breast cancer literature about the prevalence of HPV, MMTV, EBV, CMV, BLV, JCV and MCPyV for the 15-year period 2008–2023 using the keywords ((hpv) OR (human papillomavirus)) AND ((breast cancer) OR (BC) OR (breast neoplasia)) AND (pathogenesis), ((cmv)or(cytomegalovirus)) AND ((breast cancer) OR (BC) OR (breast neoplasia)) AND (pathogenesis), ((ebv)or(epstein-barr virus)) AND ((breast cancer) OR (BC) OR (breast neoplasia)) AND (pathogenesis), ((mmtv)or(mouse mammary tumor virus)) AND ((breast cancer) OR (BC) OR (breast neoplasia)) AND (pathogenesis), ((blv)or(bovine leukemia virus)) AND ((breast cancer) OR (BC) OR (breast neoplasia)) AND (pathogenesis), ((jcv)or(human polyomavirus 2)) AND ((breast cancer) OR (BC) OR (breast neoplasia)) AND (pathogenesis), ((MCPyV)or(merkel cell polyomavirus)) AND ((breast cancer) OR (BC) OR (breast neoplasia)) AND (pathogenesis).
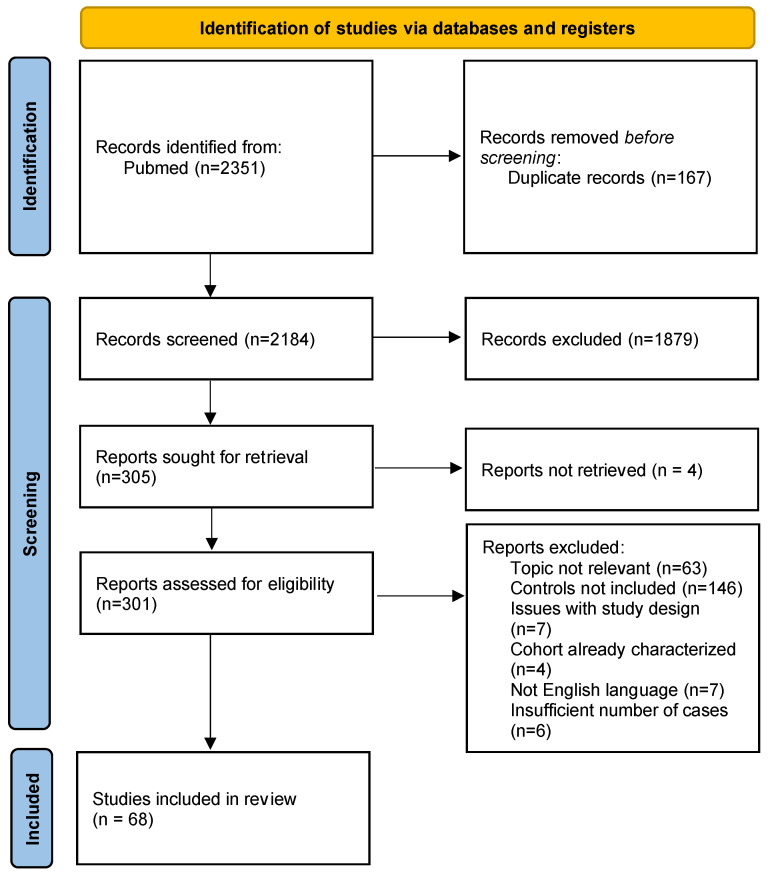
A total of 2351 publications were screened according to the 2020 Prisma statement [6]; the results are summarized in Figure 1. Articles were imported into Microsoft Excel 2021 (https://www.microsoft.com/en-us/microsoft-365/excel, accessed on 1 May 2024) and duplicates were removed. Then, title and abstracts were screened to exclude a first group of publications not relevant to this work. Then, the full texts of the remaining articles were obtained, and the articles were further screened based on their content and relevance to the topic at hand. Exclusion criteria are detailed in Figure 1. A final count of 68 articles represented our cohort, and for that, we noted the author, year of publication, number of positive cases and positive controls, and types of controls. Fisher’s exact test and Pearson’s chi-square test (GraphPad Prism 5) were used to calculate the statistical significance (p ≤ 0.05 were considered significant) of case and control prevalence when appropriate, and they are reported in the tables in the text. Since most of the articles included in the systematic review were not structured like case–control studies, we refrained from calculating the odds ratio.
Figure 1.
PRISMA 2020 flow diagram for systematic reviews.
3. HPV
HPV is a family of DNA viruses that represent the most frequent sexually transmitted disease worldwide [7]. They are able to infect both skin and genital and oral mucosa, and they are responsible for a wide range of proliferative lesions, spanning from the common cutaneous wart to invasive cancer, most notably in the uterine cervix. They are categorized into low- and high-risk (HR) genotypes based on their association with cancer development. Most infections, even with HR genotypes, are asymptomatic, do not cause any visible lesion, and are cleared by the immune system in the months following the first infection; when it is not cleared, persistent infection is the most important risk factor for the development of HPV-linked cancer. The development of effective HPV vaccines has already changed the epidemiology of cervical cancer in countries where programs have been systematically implemented [8], and the effects will become more evident also for other types of cancer as the programs progress in time and become more inclusive.
HR-HPV is linked to the development of cervical, anogenital and head and neck carcinomas; it produces the oncogenic proteins E6 and E7, which have the potential to disrupt cell cycle by promoting degradation of the oncoprotein p53 and by binding the Rb protein, respectively.
HR-HPV relevance in the pathogenesis of breast cancer has been hypothesized after the discovery that HR-HPV can immortalize mammary epithelial cells in vitro [9,10], but an actual route of infection of the ductal epithelium in vivo has not been put forward yet.
There are three hypotheses for the mechanisms of infection [11]: through blood/lymphatics in a patient with previous HPV-positive gynecological cancer, by direct contact between genital and breast during sex through skin or nipple microabrasions, or by oral transmission to breast via bodily fluids. Another hypothesis for viral carcinogenesis is the so-called ‘hit and run’ mechanism, by which a virus may be involved in starting the carcinogenesis without integration into the host genome, and then subsequently cleared by the host immune defenses [12,13]. According to this hypothesis, the absence of measurable viral genetic material does not exclude its role in initiating the cancer process, and cancers that remain positive for the virus represent only a fraction of the whole; however, at the current time, there is not sufficient evidence to favor one hypothesis over the others.
Glenn et al. [14] also reported that HPV could be identified by polymerase chain reaction (PCR) in the milk of 6/40 (15%) women with no history of breast cancer; this mechanism of transmission was reported for MMTV, in which shedding of the virus in mother’s milk and lactation significantly increased the progeny’s risk of mammary cancer. A study by the European Institute of Oncology by Cazzaniga et al. [15] reported similar rates (14%) of positivity for HPV in ductal lavages, but the positivity was mostly confined to the nipple superficial epithelium, and the authors therefore surmised that it was a colonization or transient infection in the external epithelial cells of the nipple, and could not explain spread to the ductal epithelium of the breast.
Some authors reported koilocytosis in breast cancer specimens [16,17], suggesting a possible morphological clue for the link between HPV infection and breast cancer development. Some authors also investigated the possibility that an HPV infection at another site could increase the risk of breast cancer, through spread of the virus from the primary site to the breast via blood or lymphatics, as outlined above. On this note, it has been reported that women with a previous history of cervical intraepithelial neoplasia have a higher incidence of breast cancer [18,19].
Frega et al. [20] also investigated concurrent cervical HPV infection in the nine patients whose breast cancer had tested positive for HPV in their study population. They reported six (67%) cervical infections in the tested patients; in all cases, the patient shared at least one genotype between cervical and breast positivity. In the same study, Frega et al. also investigated nodal metastasis HPV positivity, and one case showed a positive HPV16 breast primary with a positive HPV16 nodal metastasis.
Data about HPV reported prevalence in breast cancer is reported in Table 1.
Table 1.
Prevalence of HPV DNA detection in breast cancer samples according to different studies.
| Study | Positive Cases/Total Cases (%) | Genotypes Detected (Detected/Cases) | Technique Used |
|---|---|---|---|
| Haghighi, 2023 [21] | 23/90 (25%) | HPV16 (10/23), HPV18 (9/23), HPV33 (2/23). | RT-PCR on CPT |
| Maroga, 2023 [22] | 78/101 (77%) | HPV16 (37/101) HPV70 (36/101), HPV51 (22/101), HPV35 (16/101), HPV82 (16/101), HPV18 (5/101). | PCR on FFPE sections |
| Alinezhadi, 2022 [23] | 8/63 (13%) | HPV33 (3/8), HPV16 (2/8), HPV11 (1/8). | PCR on FFPE sections |
| Calderon, 2022 [24] | 13/447 (3%) | HPV16 (11/13), HPV18 (2/13). | PCR on CPT (controls) and FFPE sections (cases) |
| Gupta, 2022 [25] | 48/74 (65%) | // | PCR on FFPE TMA |
| Mamani-Zapana, 2022 [26] | 27/32 (84%) | HPV18 (19/27), HPV16 (8/27). | RT-PCR on FFPE sections |
| Oliveira, 2022 [27] | 0/54 (0%) | // | Nested-PCR on FFPE sections |
| Charostad, 2021 [28] | 12/36 (33%) | HPV16 (8/12), HPV18 (2/12), HPV31 (1/12), HPV6 (1/12). | PCR on CPT |
| Elagali, 2021 [29] | 13/150 (9%) | HPV16 (5/13), HPV58 (4/13), HPV18 (3/13), HPV11 (1/13). | PCR on FFPE sections |
| Golrokh Mofrad, 2021 [30] | 7/59 (12%) | HPV18 (5/7), HPV6 (2/7). | PCR on FFPE sections |
| Gupta, 2021 [31] | 37/70 (53%) | HPV52 (18%), HPV45 (11%), HPV31 (4%), HPV58 (4%), HPV68 (1%). | PCR on FFPE sections |
| Guo, 2021 [32] | 58/180 (32%) | HPV16/18 (39/58), HPV6/11 (19/58). | CISH on FFPE sections |
| Sher, 2020 [33] | 5/50 (10%) | HPV16 (3/5), HPV35 (3/5), HPV58 (1/5). | PCR on fresh breast tissue |
| Tawfeik, 2020 [34] | 4/20 (20%) | HPV16 (3/4), HPV18 (1/4). | RT-PCR on CPT |
| Balci, 2019 [35] | 8/18 (44%) | HPV11 (8/8), HPV39 (2/8), HPV6 (1/8), HPV82 (1/8) | PCR on FFPE sections |
| Baltzell, 2018 [36] | 2/61 (3%) by IS-PCR 4/61 (7%) by ISH |
HPV16 | IS-PCR and ISH on FFPE sections |
| Cavalcante, 2018 [37] | 51/103 (50%) | HPVX (40/51), HPV6/11 (7/51), HPV18 (2/51), HPV33 (2/51) | Nested-PCR on FFPE samples |
| Ghaffari, 2018 [38] | 4/72 (6%) | // | Nested-PCR on FFPE sections |
| Habyarimana, 2018 [39] | 22/47 (47%) | HPV16 (77%), HPV33(14%), HPV31 (9%) | Nested-PCR on FFPE sections |
| Kouloura, 2018 [40] | 0/201 (0%) | // | E1-based PCR on TMA |
| Malekpour Afshar, 2018 [41] | 8/98 (8%) | HPV16 (5/8), HPV18 (5/8), HPV31 (3/8), HPV33 (3/8) | rtPCR on FFPE |
| Bakhtiyrizadeh, 2017 [42] | 0/150 (0%) | // | PCR on FFPE |
| Delgado-García, 2017 [43] | 130/251 (52%) | // | PCR on FFPE sections |
| Islam, 2017 [44] | 174/272 (64%) | HPV16 (120/174), HPV18 (61/174) and HPV33 (5/174). | PCR and IHC on fresh tissue |
| Naushad, 2017 [45] | 45/250 (18%) | PCR on FFPE sections | |
| Salman, 2017 [46] | 35/74 (47%) | HPV39 (13/35), HPV18 (8/35), HPV45 (8/35), HPV16 (7/35), HPV35 (7/35), HPV59 (7/35). | type-specific PCR on fresh tissue |
| Wang, 2017 [47] | 14/81 (17%) | // | hybrid capture 2 on fresh tissue |
| Wang, 2017 [48] | 14/50 (28%) | HPV16 (14/14) | qRT-PCR on FFPE |
| Doosti, 2016 [49] | 20/87 (23%) | HPV6 (9/20), HPV16 (7/20), HPV18 (3/20), HPV11 (1/20). | Nested-PCR on FFPE |
| Ilahi, 2016 [50] | 8/46 (17%) | HPV16 (8/8) | PCR on FFPE sections |
| Yan, 2016 [51] | 23/76 (30%) | HPV18 (23/23) | Dual-PCR |
| Wang, 2016 [52] | 52/146 (36%) | ISH on FFPE sections | |
| Li, 2015 [53] | 3/187 (2%) | HPV6 (1/3), HPV18 (1/3), HPV16 (1/3). | PCR on FFPE |
| Fimereli, 2015 [54] | 2/58 (3%) with PCR 0/58 (0%) with transcriptome |
HPV16 (1/2), HPV-HR (1/2) | RNA sequencing, exome sequencing PCR and IHC on CPT and FFPE sections |
| Fu, 2015 [55] | 25/169 (15%) with PCR 17/169 (10%) with ISH |
HPV58 (25/25) | PCR and ISH on FFPE |
| Gannon, 2015 [56] | 13/80 (16%) | Nested-PCR on CPT and RNA sequencing | |
| Salehpour, 2015 [57] | 54/206 (26%) | HPV11 (19/54), HPV6 (5/54), HPVX (48/54). | PCR on FFPE sections |
| Vernet-Tomas, 2015 [58] | 0/76 (0%) | // | PCR on FFPE sections |
| Ahangar-Oskouee, 2014 [59] | 22/65 (34%) | HPV6 (17/22), HPV11 (1/22), HPV16 (1/22), HPV35 (1/22), HPV52 (1/22). | Nested-PCR on FFPE |
| Ali, 2014 [60] | 60/129 (47%) | HPV31 (39/129), HPV18 (35/129), HPV16 (33/129), HPV33 (16/129). | ISH on FFPE sections |
| Corbex, 2014 [61] | 15/123 (12%) | HPV16 (8/15), HPV31 (3/15), HPV22 (2/15). | PCR on FFPE sections |
| Manzouri, 2014 [62] | 10/55 (18%) | HPV12 (2/10), HPV11 (2/10), HPV8 (1/10), HPV33 (1/10), HPV35 (1/10), HPV45 (1/10), HPV55 (1/10). | PCR on FFPE sections |
| Peng, 2014 [63] | 2/100 (2%) | HPV18 (2/2) | Multiplex PCR detected by matrix-assisted laser desorption ionization-time of flight mass spectrometry on CPT |
| Piana, 2014 [64] | 6/80 (8%) | HPV16 (28.6%), HPV31 (14.3%), HPV45 (14.3%), HPV52 (14.3%), HPV6 (14.3%), HPV66 (14.3%) | PCR on FFPE sections |
| Herrera-Goepfert, 2013 [65] | 8/20 (40%) | HPV16 (7/8), HPV18 (1/8). | PCR on FFPE sections |
| Hossein, 2013 [66] | 52/150 (35%) | HPV16 (21/52), HPV18 (15/52), HPV11 (8/52), HPV31 (4/52), HPV33 (2/52), HPV35 (2/52). | PCR on FFPE sections |
| Lieng, 2013 [67] | 48/224 (21%) | // | hybrid capture 2 assay |
| Pereira Suarez, 2013 [68] | 16/61 (26%) | HPV11 (4/16), HPV16 (3/16), HPV13 (1/16). | PCR on fresh tissue |
| Baltzell, 2012 [69] | 2/70 (3%) with IS-PCR 4/70 (6%) with ISH |
// | ISH and IS-PCR on FFPE sections |
| Chang, 2012 [70] | 0/48 (0%) | // | FQ-PCR and ISH on FFPE sections |
| Frega, 2012 [20] | 9/31 (29%) | HPV16 (44%), HPV6 (22%). | // |
| Glenn, 2012 [17] | 25/50 (50%) with liquid PCR 10/27 (37%) with IS-PCR |
HPV18 (25/25) | Liquid PCR on CPT, IS-PCR on FFPE sections |
| Herrera-Romano, 2012 [71] | 0/118 (0%) | // | PCR on FFPE microdissected FFPE sections |
| Sigaroodi, 2012 [72] | 15/79 (26%) | HPV18 (4/15), HPV16 (4/15), HPV6 (2/15), HPV23 (2/15) HPV124 (1/15), HPV15 (1/15), HPV11 (1/15). | PCR on FFPE sections |
| Aguayo, 2011 [73] | 4/46 (9%) | HPV16 (4/4) | RT-PCR on FFPE sections |
| Antonsson, 2011 [74] | 27/54 (50%) | HPV18 (27/27) | PCR on CPT and ISH on FFPE |
| Ghaffari, 2011 [75] | 30% (out of 67; exact number not provided) | HPV16 (15%). HPV11/16 (12%), HPV31/33 (3%). Exact number not provided | PCR on CPT |
| Hedau, 2011 [76] | 0/228 (0%) | // | qRT-PCR on CPT |
| Mou, 2011 [77] | 4/62 (7%) | HPV16 (3/4), HPV18 (1/4) | PCR on CPT |
| Silva, 2011 [78] | 0/79 (0%) | // | PCR on FFPE sections |
| Hachana, 2010 [79] | 0/123 (0%) | // | PCR and ISH on FFPE sections |
| Yavuzer, 2010 [80] | 0/84 (0%) | // | Nested-PCR on FFPE sections |
| De León, 2009 [81] | 15/51 (29%) | HPV16 (11/15), HPV18 (4/15). | PCR on FFPE sections |
| He, 2009 [82] | 24/40 (60%) | HPV16 (24/24). | PCR on CPT |
| Heng, 2009 [16] | 8/26 (31%) | HPV18 (7/8), HPV16 (1/8). | IS-PCR on FFPE |
| Mendizabal-Ruiz, 2009 [83] | 3/67 (4%) | HPV31 (2/3), HPV6 (1/3), HPV16 (1/3), HPV35 (1/3), HPV18 (1/3) | PCR on FFPE |
| Akil, 2008 [10] | 69/113 (61%) | HPV33 (63/69), HPV35 (42/69), HPV18 (11/69), HPV16 (10/69), HPV31 (8/69) | PCR on FFPE TMA |
| de Cremoux, 2008 [84] | 0/50 (0%) | // | PCR on unspecified material |
| Duò, 2008 [85] | 2/52 (4%) | HPV66, HPVX | PCR on FFPE sections |
| Khan, 2008 [86] | 26/124 (21%) | HPV16 (24/26), HPV6 in (12/26), HPV18 (3/26), HPV51 (2/26), HPV33 (1/26). | qRT-PCR on FFPE |
RT-PCR, real-time PCR; CPT, cryopreserved tissue; //, data not available; FFPE, formalin-fixed, paraffin-embedded; TMA, tissue microarray; CISH, chromogenic in situ hybridization; IS-PCR, in situ PCR; qRT-PCR, quantitative real-time PCR; ISH, in situ hybridization; IHC, immunohistochemistry; FQ-PCR, fluorescence quantitative PCR.
Reported prevalence for HPV ranged from 0% to 84% in the indexed literature in our study timeframe, with a median value of 18%, as reported in Table 1. Coinfection was frequently reported [10,20,22,26,31,35,43,44,52,57,60,64,68,72,81,83,86]; while coinfection has been found to be associated with higher stage and higher grade in cervical HPV-related lesions, no significant association has been reported in breast cancer in any of the studies reporting on it.
Variation in the reported prevalence can be due to a number of factors: choice of primer regions for PCR amplification, the sensibility of the techniques used for retrieval and amplification of viral DNA, source material (FFPE or CPT), storing conditions of the sample. Some of the studies employed a restricted set of HPV primers and only investigated HR genotypes, while others also screened for low-risk genotypes. Also, geographical variation of HPV distribution in the population is a known finding in cervical lesions, and it is also reflected in the diverse prevalence of HPV genotypes in different populations, and therefore may be reflected in a different prevalence also in breast cancer.
While it is difficult to compare the prevalence of specific HPV genotypes, due to the large variation of HPV primers, recent systematic reviews report the high-risk HPV16 as the most commonly detected HPV genotype in breast cancer [87,88]; other commonly reported genotypes are HPV18 and HPV33, both high-risk genotypes. From Table 1, our systematic review of the literature confirms HPV16, HPV18 and HPV33 as the most commonly detected HPV genotypes; however, we feel that these results are too dependent on the primer choice to be able to make strong inferences from them in this setting. Studies with a larger set of primers have also identified low-risk genotypes such as HPV6, whose relevance for breast carcinogenesis is unclear.
Choice of identification technique also has a significant impact on the results, and should be kept in mind when critically examining the results of the studies and comparing the reported incidence of HPV DNA in breast cancer [89]. Most of the studies use PCR to amplify the viral genome, a technique that is prone to contamination; it also makes it impossible, after amplification, to detect if the positivity for HPV comes from the breast cancer cells or from bystander stromal or immune cells. This is of particular relevance not only for HPV, but also for other viruses, such as EBV, that is often detected in lymphocytes.
When studies compared PCR rate of viral detection with in situ techniques, the reported positivity was lower in the in situ techniques rather than PCR [17,55]. Antonsson et al. [74] reported a 50% positivity for HPV when testing for the virus on CPT with PCR, but failed to report any positivity in tumor or adjacent breast tissue with ISH on FFPE. Frega et al. [20] reported nine positive cases on FFPE, but failed to demonstrate any viral mRNA positivity. Gannon et al. [56] sequenced the RNA transcript of 5 breast cancer samples that had tested positive for HPV DNA with PCR, as well as 53 available breast cancer and 10 normal breast tissue datasets, and did not identify any viral transcript.
This difference in the detection rate between PCR and ISH [55] has been explained by some authors as due to a low viral load in the ISH−/PCR+ cases; others consider this a possible spurious amplification of passerby or non-cancer related HPV genome by PCR. In general, HPV concentration in breast cancer seems to be lower than the concentrations detected in cervical cancer, making its detection more difficult [90].
Another important factor to keep in mind is that most of the studies reported only the detection of HPV DNA in the cancer tissue, without evaluating the presence of E6 and E7 transcripts, which is a hallmark of functional activity of the virus; therefore, we are not able to comment if the detected virus was actually able to influence the cellular milieu or was just an incidental finding.
Aguayo et al. [73] investigated both the integration and functionality status of HPV, and found the virus to be integrated and transcribed, albeit with a low viral load, but not functional.
Guo et al. [32], on the other hand, investigated the presence of HPV by chromogenic in situ hybridization (CISH), and only reported the CISH as positive when an integrative pattern was present; they therefore reported a 32% of breast cancer patients (both in situ and invasive) who were positive for integrated HPV DNA. In the same article, they also found HR-HPV to be significantly more amplified in the cancer than in the surrounding normal breast tissue [32]; no correlation with survival or clinico-pathological characteristics of the patients was found.
The study by Islam et al. [44] reported that 88% of HPV16 was present as integrated into the DNA of breast cancer. They also reported an increased number of HPV16 copies with increasing histological grade, stage and lymph node metastasis, and presence of the E6/E7 transcripts, indicating the function of the viral integration.
Khan et al. [86] reported the presence of integrated HPV DNA in all their 24 HPV16-positive cases; they did not report any instance of episomal DNA. Wang et al. [52] reported the presence of both episomal and integrated viral DNA in breast cancer; they did not, however, detect any significant viral transcription.
Due to the significant variation in conditions, choice of primers and population, it is not an easy task to compare results across the studies, also regarding the characteristics of breast cancer that different authors identified as possibly linked to HPV infection.
Most of the studies included cases that were a mix of invasive breast cancer histological types, with ductal carcinoma/carcinoma of no special type being the most commonly reported.
Some papers focused on specific histological types or molecular subtypes. Piana et al., Gupta et al. and Corbex et al. [31,61,64] investigated triple negative breast cancer in an Italian, a Croatian and an Algerian population. In their study, Piana et al. [64] reported a 15% vs. 0% of HPV positivity in triple negative vs. non-triple negative breast cancer. Corbex et al. [61] did report that triple negative breast cancer and inflammatory breast cancer had a stronger association to viral detection than other histological types, but did not link any specific virus to either triple negative or inflammatory breast cancer.
Herrera-Goepfert et al. [65] investigated HPV positivity rates in metaplastic breast cancer, and reported higher copy number than usually found in other histological types, while Duò et al. [85] performed PCR on 52 cases of breast cancer and found two positive results; both were invasive papillary carcinoma. Lastly, one case report by Kulka et al. [91] reported detection of HPV18 and HPV33 in a lymphoepithelioma-like carcinoma of the breast.
Islam et al. [44] reported worse survival in patients with breast cancer with positive HPV-DNA in tissue with respect to HPV-negative breast cancer patients, but it is the only study to report this association. Most studies [45,47,55,65,67,68,86] do not report any difference in age, tumor size, lymph node status or histological prognostic and predictive factors. De León et al. [81] reported a correlation with HPV DNA positivity and larger tumor size, but Antonsson et al. and Ghaffari et al. [74,75] reported the opposite, with HPV-positive tumors being smaller than HPV-negative counterparts, and having less nodal involvement.
Therefore, we do not consider HPV DNA detection in breast cancer to be associated with any difference in prognosis or tumor characteristics with respect to HPV-negative cancer.
To complete our review of the relevance of HPV-positivity in breast cancer, we analyzed the selected articles to evaluate the difference in prevalence between breast cancer and controls. The data are summarized in Table 2, as follows.
Table 2.
Summary of the prevalence of HPV detection of investigated cases and controls.
| Study | HPV-Positive Cases/Total | HPV-Positive Controls/Total | Statistically Significant? Y/N (p-Value) |
|---|---|---|---|
| Haghighi, 2023 [21] | 23/90 (25%) | 7/32 (22%) ^ | No (p = 0.8127) |
| Alinezhadi, 2022 [23] | 8/63 (13%) | 9/32 (28%) * | No (p = 0.0890) |
| Calderon, 2022 [24] | 5/447 (3%) | 1/79 (1%) # | No (p = 1.0000) |
| Golrokh Mofrad, 2021 [30] | 7/59 (12%) | 0/11 (0%) # | No (p = 0.5866) |
| Charostad, 2021 [28] | 12/36 (33%) | 2/36 (6%) § | Yes (p = 0.0059) |
| Guo, 2021 [32] | 58/180 (32%) | 10/131 (8%) ^,$ | Yes (p < 0.0001) |
| Sher, 2020 [33] | 5/50 (10%) | 8/100 (8%) *,#,$ | No (p = 0.7607) |
| Tawfeik, 2020 [34] | 4/20 (20%) | 0/15 (0%) # | No (p = 0.1186) |
| Baltzell, 2018 [36] | 2/61 (3%) by IS-PCR 4/60 (7%) by ISH |
8/100 (8%) § by IS-PCR 3/95 (3%) by ISH |
No by IS-PCR (p = 0.3211) No by ISH (p = 0.4310) |
| Cavalcante, 2018 [37] | 51/103 (50%) | 15/95 (16%) ^ | Yes (p < 0.0001) |
| Malekpour Afshar, 2018 [41] | 8/98 (8%) | 0/40 & | No (p = 0.1046) |
| Bakhtiyrizadeh, 2017 [42] | 0/150 (0%) | 0/150 # | // |
| Delgado-García, 2017 [43] | 130/251 (52%) | 49/186 (26%) # | Yes (p < 0.0001) |
| Islam, 2017 [44] | 174/272 (64%) | 10/38 (26%) ^,* | Yes (p < 0.0001) |
| Salman, 2017 [46] | 35/74 (47%) | 11/36 (31%) ^,# | No (p = 0.1045) |
| Doosti, 2016 [49] | 20/87 (23%) | 0/84 & | Yes (p < 0.0001) |
| Wang, 2016 [52] | 52/146 (35%) | 3/83 # | Yes (p < 0.0001) |
| Li, 2015 [53] | 3/187 (2%) | 0/92 # | No (p = 0.5532) |
| Gannon, 2015 [56] | 13/80 (16%) | 1/10 (10%) # | No (p = 1.0000) |
| Ahangar-Oskouee, 2014 [59] | 22/65 (34%) | 0/65 *,#,$ | Yes (p < 0.0001) |
| Ali, 2014 [60] | 60/129 (47%) | 0/20 (0%) ^ | Yes (p < 0.0001) |
| Manzouri, 2014 [62] | 10/55 (18%) | 7/51 (14%) # | No (p = 0.6029) |
| Peng, 2014 [63] | 2/100 (2%) | 0/100 # | No (p = 0.4975) |
| Lieng, 2013 [67] | 48/224 (21%) | 6/37 (16%) * | No (p = 0.6611) |
| Chang, 2012 [70] | 0/48 (0%) | 3/30 (10%) # | No (p = 0.0534) |
| Frega, 2012 [20] | 9/31 (29%) | 0/12 *,$ | Yes (p = 0.0438) |
| Glenn, 2012 [17] | 10/27 (37%) | 3/18 (17%) ^ | No (p = 0.1882) |
| Sigaroodi, 2012 [72] | 15/79 (26%) | 1/41 (2%) § | Yes (p = 0.0105) |
| Mou, 2011 [77] | 4/62 (7%) | 0/46 (0%) § | No (p = 0.1345) |
| de León, 2009 [81] | 15/51 (29%) | 0/43 (0%) *,& | Yes (p < 0.0001) |
| He, 2009 [82] | 24/40 (60%) | 1/20 (5%) ^ | Yes (p < 0.0001) |
| Heng, 2009 [16] | 8/26 (31%) | 3/17 (18%) ^ | No (p = 0.4801) |
| Mendizabal-Ruiz, 2009 [83] | 3/67 (4%) | 0/40 (0%) # | No (p = 0.2911) |
* fibroadenomas, # unspecified benign lesions, § normal breast tissue from the same patient, ^ normal breast tissue from healthy patients, $ breast papillomas, & fibrocystic disorder, //, data not available.
Pooling cases and controls together, there is an HPV prevalence of 24% (840/3450) in the cases and of 8% (158/1890) controls. p-value was significant (p < 0.0001).
However, it is not so straightforward to compare results across different papers, and therefore, the statistical power of this pooled analysis must also be taken with caution.
Study design between different papers is extremely heterogeneous, especially in regard to the control selection; in this systematic review, 7 control groups were fibroadenomas, 15 were unspecified benign lesions, 5 were normal breast tissue from the same patient, 8 were normal breast tissue from healthy patients, 4 were breast papillomas, 3 were fibrocystic disorder, and 6 studies included more than one of these categories. Chang et al. [70] reported HPV positivity in three benign phyllodes tumors. On the same topic of HPV positivity in fibroepithelial tumors, Alinezhadi et al. [23] also reported a 28% of positivity in their fibroadenoma control group, higher than the positivity in the control group; they also reported that this group had a lower incidence of HR-HPV genotypes with respect with the positives in the case group. While this may indicate that different HPV genotypes in breast may cause a spectrum of diseases, not only limited to epithelial malignancies or benign changes, it also raises issues with the inclusion of fibroepithelial tumors as negative controls in some of the case–control studies. Two studies from Gupta et al. [31] and Piana et al. [64] were excluded because the control group included malignant cases, to minimize the variability between control groups for the sake of this review.
The use of different controls did not impact the significance of the study. Since very different populations are used as controls in different papers, some variation would be expected; the fact that it is not observed is probably linked to a deficiency in statistical power from different studies, rather than an actual homogeneity.
Therefore, even if the pooled analysis is strongly significant, and this might imply some influence of HPV infection on breast cancer, we feel that the limitations in the design of the different studies detailed in this paragraph must be considered, and caution should be taken when analyzing the relevance of these data.
4. MMTV
Mouse mammary tumor virus (MMTV) is a betaretrovirus known to cause mammary cancer and lymphoma in mice, and has been identified also in breast milk. It was first discovered in 1936 by Bitter et al. [2] as a cancer-causing agent secreted in breast milk, and it sparked the notion that viruses can be the etiological agent of cancer. A human analogous virus, human mammary tumor virus (HMTV) or MMTV-like virus, has been proposed as a causative agent for human breast cancer [92].
However, in situ hybridization studies demonstrated that the detected DNA was homologous to human endogenous retroviruses [93] that represent vestigia from remote infections that were integrated into human DNA and have become inactivated by millennia of mutations. Moreover, the fact that there is no known cell receptor for MMTV in human cells also has not been explained or demonstrated in vivo, even if groups have reported transmission in in vitro cell cultures [94].
Correlation of the geographical incidence of breast cancer with the distribution of different mouse species in the early 2000s has been made [95], with Mus domesticus, the common house mouse, roughly paralleling the frequency of MMTV detection in breast cancer.
A study [96] by a group from University of Pisa reported the presence of MMTV DNA and RNA in saliva of children and adults, and in higher percentage in breast cancer patients, and proposed saliva as a route of interpersonal spread of infection. The presence of MMTV env genes have also been reported in 4% of human milk samples from healthy lactating women [97].
Data about MMTV reported prevalence in breast cancer is reported in Table 3.
Table 3.
Prevalence of MMTV detection in breast cancer samples according to different studies.
| Study | Positive Cases/Total Cases (%) | Technique Used |
|---|---|---|
| Fekete, 2023 [98] | 0/75 (0%) | PCR for MMTV-like env gene on FFPE sections |
| Gupta, 2022 [25] | 11/74 (15%) | PCR on FFPE sections |
| Gupta, 2021 [31] | 5/70 (7%) | PCR on FFPE sections |
| Khalid, 2021 [99] | 69/105 (65%) | qPCR on FFPE sections |
| De Sousa Pereira, 2020 [100] | 41/217 (19%) | Nested-PCR for MMTV-like env gene on fresh tissue |
| Naccarato, 2019 [101] | 19/103 (18%) | Fluorescence nested-PCR of microdissected FFPE sections for MMTV env-like sequences gene on fresh tissue |
| Seo, 2019 [102] | 12/128 (9%) | PCR for HMTV on fresh tissue |
| Al Dossary, 2018 [103] | 9/103 (9%) | Reverse transcriptase PCR for MMTV env-like sequences on FFPE sections |
| Motamedifar, 2017 [104] | 0/50 (0%) | PCR for MMTV env genes on FFPE sections |
| Naushad, 2017 [45] | 83/250 (29%) | PCR on FFPE sections |
| Shariatpanahi, 2017 [105] | 19/59 (32%) | Nested-PCR for MMTV-like env sequences on FFPE sections |
| Reza, 2015 [106] | 12/100 (12%) | RT-PCR of FFPE sections |
| Ahangar Oskouee, 2014 [107] | 0/65 (0%) | Nested-PCR for MMTV-like sequences on FFPE sections |
| Naushad, 2014 [108] | 16/80 (20%) for env gene 21/80 (26%) for LTR sequences |
PCR for MMTV env-like and LTR sequences on FFPE sections |
| Morales-Sánchez, 2013 [109] | 0/86 (0%) | PCR for env gene on CPT |
| Tabriz, 2013 [110] | 0/40 (0%) | RT-PCR for MMTV-like env gene on FFPE sections |
| Glenn, 2012 [17] | 39/50 (78%) with liquid PCR, 5/27 (19%) with IS-PCR |
Liquid PCR on fresh frozen tissue, IS-PCR on FFPE sections |
| Mazzanti, 2011 [111] | 7/20 (35%) | Fluorescence nested-PCR for MMTV env-like sequences on microdissected FFPE sections |
| Park, 2011 [112] | 0/42 (0%) | Nested-PCR for MMTV-like env sequences on FFPE sections |
| Lawson, 2010 [113] | 33/74 (54%) by PCR 5/27 (19%) by IS-PCR |
IS-PCR and IHC for the gp52 env protein on FFPE sections |
| Fukuoka, 2008 [114] | 0/46 (0%) | PCR for MMTV env sequences and Southern blot hybridization on CPT |
| Hachana, 2008 [115] | 17/122 (14%) | Nested-PCR for MMTV env like on CPT |
FFPE, formalin-fixed, paraffin-embedded; qPCR, quantitative PCR; RT-PCR, real-time PCR; CPT, cryopreserved tissue; IS-PCR, in situ PCR; IHC, immunohistochemistry.
Reported prevalence for MMTV ranged from 0% to 78% in the indexed literature in our study timeframe, with a median value of 15%, as reported in Table 3. As in the previous section, we excluded a paper by Cedro-Tanda et al. [116] because the cases included both benign and malignant lesions, and epithelial and fibroepithelial lesions; the decision was taken to keep the cohort of the review as homogeneous as possible as to minimize confounding factors.
One of the limits of interpretation of these data is that the only technique that is used for the detection of the virus is PCR, which is, as previously mentioned, a technique with several limits regarding localization of the virus positivity and extremely prone to contamination. Al Dossary et al. [103] actually reported a higher prevalence of MMTV-like env sequences in the adjacent normal tissue than in the cancer cells itself (10% vs. 6%). Perzova et al. [117] reported very low viral copy numbers in the analyzed samples, and deemed this result incompatible with a retrovirus-caused cancer; in their report, contamination from mice infesting the building was the most likely source of the unconvincing results.
One issue that has been raised by some authors as to explain the absence of viral transcripts in some of the experiments is the poor specificity of the PCR primer for the env sequence. However, the work by Fukuoka et al. [114] utilized the same primers as Pogo et al. [118] and still reported an absence of detection of virus in the 49 breast cancer cases sequenced.
Regarding integration of the virus in breast cancer cells, Lawson et al. [113] reported detection of the gp52 env protein by IHC in the nucleus of the cell; they claim this as proof of integration of the viral DNA into the cell after infection. It must be noted, however, that cross-reactivity has been documented in the literature [119,120].
Lawson et al. [113] also reported that the MMTV-positive breast cancer had ‘very similar’ histological features to the MMTV-positive mouse mammary tumors, with the description given in the paper resembling a medullary-like phenotype of human breast cancer. MMTV infection has been postulated to be linked to the development of pregnancy-associated breast cancer [121] and of inflammatory breast cancer [122], but not in a definitive manner.
Several authors [25,100,102,108,123] failed to report any statistical association between MMTV-like virus and clinicopathological characteristics of breast cancer. Hachana et al. [115] reported an inverse correlation between MMTV presence and progesterone receptor and HER2 status. De Sousa Pereira et al. [100] reported an increased frequency of MMTV-like env gene in HER2-enriched breast cancer, both of non-luminal and luminal histology. Naccarato et al. [101] found that MMTV env-like sequences were significantly more present in sporadic rather than hereditary breast cancer (30% vs. 4%). Mazzanti et al. [111] also evaluated the prevalence of MMTV in pre-invasive lesions and reported it in 27% of atypical ductal hyperplasia (ADH) and 82% of ductal carcinoma in situ.
To complete our review of the relevance of MMTV positivity in breast cancer, we analyzed the selected articles to evaluate the difference in prevalence between breast cancer and controls. The data are summarized in Table 4, as follows.
Table 4.
Summary of the prevalence of MMTV detection of investigated cases and controls.
| Study | MMTV-Positive Cases/Total | MMTV-Positive Controls/Total | Statistically Significant? Y/N (p-Value) |
|---|---|---|---|
| Khalid, 2021 [99] | 69/105 (65%) | 2/15 (13%) § | Yes (p = 0.0001) |
| De Sousa Pereira, 2020 [100] | 41/217 (19%) | 30/196 (15%) § | No (p = 0.3624) |
| Seo, 2019 [102] | 12/128 (9%) | 0/128 (0%) § | Yes (p = 0.0004) |
| Al Dossary, 2018 [103] | 9/103 (9%) | 0/51 *,#,& | Yes (p = 0.0301) |
| Shariatpanahi, 2017 [105] | 19/59 (32%) | 3/59 (5%) # | Yes (p < 0.001) |
| Reza, 2015 [106] | 12/100 (12%) | 0/100 (0%) § | Yes (p = 0.0003) |
| Ahangar Oskouee, 2014 [107] | 0/65 (0%) | 0/65 # | // |
| Morales-Sánchez, 2013 [109] | 0/86 (0%) | 0/65 (0%) § | // |
| Glenn, 2012 [17] | 39/50 (78%) with liquid PCR 5/27 (19%) with IS-PCR |
13/40 (32%) ^ with liquid PCR 3/18 (17%) ^ with IS-PCR |
Yes, with liquid PCR (p = 0.0001) No with IS-PCR (p = 1.0000) |
| Lawson, 2010 [113] | 33/74 (54%) with liquid PCR 5/27 (19%) by IS-PCR |
0/29 (0%) § with liquid PCR 3/18 (17%) § with IS-PCR |
Yes with liquid PCR (p = 0.0001) No with IS-PCR (p = 1.0000) |
| Hachana, 2008 [115] | 17/122 (14%) | 0/122 (0%) § | Yes (p < 0.0001) |
* fibroadenomas, # unspecified benign lesions, § normal breast tissue from the same patient, ^ normal breast tissue from healthy patients, & fibrocystic disorder, //, data not available.
Pooling cases and controls together, there is an MMTV prevalence of 18% (189/1039) in the cases and of 5% (41/837) controls. p-value was significant (p < 0.0001).
Regarding the study design, there was a marked prevalence of normal breast tissue from the case group patients (7/11 studies) as control samples; while this is interesting regarding the restriction of detection of the virus to the tumor itself, it is not the common design of a case–control study, and therefore limits the strength of any consideration that could be made regarding epidemiological impact of MMTV infection.
For MMTV infection, the pooled analysis is strongly significant; however, the numbers and the design of the studies lack epidemiological power, even if some of the evidence might be compelling in suggesting a possible link.
5. EBV
EBV is a common herpesvirus in humans that infects B lymphocytes, and it is the etiological agent of infectious mononucleosis. It is estimated that close to 95% of adults have been infected worldwide, with infection rate peaking in late adolescence or earlier in developing countries. As with other herpesviruses, after the first infection, it is able to persist in the body in a latent stage, eventually recurring upon triggering. The primary route of transmission is oral, via the sharing of saliva of an infected individual, by infection of the epithelial cells of the tonsils; from there, the virus is able to infect B lymphocytes, and move to the germinal centers where the lymphocytes differentiate into memory B lymphocytes, where the virus reside in its latent phase [124,125]. It is strongly associated with nasopharyngeal and gastric cancer, endemic Burkitt’s lymphoma, post-transplant lymphoproliferative disorders and a proportion of Hodgkin’s lymphoma. Given the high prevalence of EBV antibodies in the normal population, and the fact that just a minority of people carrying a latent EBV infection develop cancer, establishing linkage and mechanisms of oncogenesis is not easy.
The possibility of a ‘hit and run’ mechanism (previously mentioned in the HPV section) has been suggested to explain a possible role of EBV in carcinogenesis for breast cancer. Some authors have reported that breast cancer cell lines infected with EBV have dysregulated HER2 and HER3 pathways [126]; other authors hypothesize that EBV infection may not be a causative agent, but rather infection during the course of the disease may alter the phenotype of the cancer, conferring a worse overall prognosis. Glenn et al. [14] reported that EBV could be identified by PCR in the milk of 14/40 (33%) women with no history of breast cancer.
Lorenzetti et al. [127] reported the detection in breast cancer cells of LMP2A, a gene associated with latency pattern II and III, that is reported in EBV-related neoplasms such as Hodgkin’s lymphoma and post-transplant lymphoproliferative disorders.
Data about EBV reported prevalence in breast cancer is reported in Table 5.
Table 5.
Prevalence of EBV detection in breast cancer samples according to different studies.
| Study | Positive Cases/Total Cases (%) | Viral Proteins Detected | Technique Used |
|---|---|---|---|
| Gouadfel, 2023 [128] | 7/30 (23%) | LMP1 | IHC on FFPE sections |
| Mekrazi, 2023 [129] | 0/100 (0%) with IHC 44/100 (44%) with PCR 0/15 (0%) with ISH |
// | IHC, PCR, ISH on FFPE sections |
| Gupta, 2022 [25] | 36/74 (49%) | // | PCR on FFPE sections |
| Oliveira, 2022 [27] | 5/72 (7%) with ISH 50/72 (69%) with IHC |
EBNA1, EBER1 | ISH and IHC on FFPE sections |
| Shahi, 2022 [130] | 25/130 (19%) | // | PCR and IHC on FFPE |
| Zhang, 2022 [131] | 57/140 (41%) | // | CISH on FFPE sections |
| Ghaffari, 2021 [132] | 12/72 (17%) | // | PCR on FFPE sections |
| Gupta, 2021 [31] | 25/70 (36%) | EBNA1 (55/70), EBNA2 (31/70) | PCR on FFPE sections |
| Charostad, 2021 [133] | 6/51 (12%) | // | PCR on CPT |
| Golrokh Mofrad, 2020 [134] | 4/59 (7%) | EBNA 1 | PCR on FFPE |
| Mostafaei, 2020 [135] | 38/83 (46%) | // | PCR on unspecified tissue sample |
| Dowran, 2019 [136] | 0/150 (0%) | // | PCR on FFPE sections |
| Sharifpour, 2019 [137] | 10/37 (27%) | EBNA3C | PCR on FFPE sections |
| Pai, 2018 [138] | 25/83 (30%) | // | ISH on FFPE slides |
| El-Naby, 2017 [139] | 10/42 (24%) | EBNA1, LMP1 | Nested-PCR on IHC on FFPE slides |
| Naushad, 2017 [45] | 83/250 (29%) | EBNA2 | PCR on FFPE sections |
| Aboulkassim, 2015 [140] | 56/108 (52%) | LMP1 EBNA1 | PCR on TMA |
| Ballard, 2015 [141] | 63/160 (39%) | // | IHC on TMA |
| Fimereli, 2015 [54] | 1/58 (1%) with transcriptome, 0/58 (0%) with IHC |
RNA sequencing, exome sequencing PCR and IHC on CPT and FFPE sections | |
| Reza, 2015 [106] | 8/100 (8%) with rt-PCR, 18/100 (18%) EBER RNA |
// | RT-PCR |
| Richardson, 2015 [142] | 24/70 (34%) | // | qPCR on CPT |
| Corbex, 2014 [61] | 10/123 (8%) | EBV1 gene | PCR on FFPE sections |
| Peng, 2014 [63] | 60/100 (60%) | // | Multiplex PCR detected by matrix-assisted laser desorption ionization-time of flight mass spectrometry on CPT |
| Marrão, 2014 [143] | 22/85 (26%) | // | RT-quantitative light cycler PCR |
| Morales-Sánchez, 2013 [109] | 4/86 (5%) | // | Nested-PCR on CPT |
| Baltzell, 2012 [144] | 2/70 (3%) with PCR, 0/70 (0%) with ISH |
// | ISH and IS-PCR on FFPE |
| Glenn, 2012 [17] | 34/50 (68%) with liquid PCR, 5/27 (19%) with IS-PCR |
EBNA1 | Liquid PCR on CPT, IS-PCR on FFPE sections |
| Zekri, 2012 [145] | 32/90 (35%) | EBNA1 EBER1 | PCR and ISH on FFPE slides |
| Aguayo, 2011 [73] | 3/46 (7%) | // | RT-PCR and ISH on FFPE sections |
| Hachana, 2011 [146] | 33/123 (27%) with PCR 0/123 (0%) with ISH 0/123 (0%) with IHC |
// | PCR, ISH and IHC on FFPE sections |
| Kadivar, 2011 [147] | 0/100 (0%) | // | IHC and PCR on FFPE sections |
| Mazouni, 2011 [148] | 65/196 (33%) | // | RT-PCR on CPT |
| Lorenzetti, 2010 [127] | 22/71 (31%) | // | ISH and PCR on FFPE |
| Joshi, 2009 [149] | 28/58 (55%) | EBNA 1 | IHC on FFPE slides |
| Fawzy, 2008 [150] | 10/40 (25%) | // | IHC on FFPE sections |
IHC, immunohistochemistry; FFPE, formalin-fixed, paraffin-embedded; //, data not available; ISH, in situ hybridization; CISH, chromogenic in situ hybridization; TMA, tissue microarray; CPT, cryopreserved tissue; RT-PCR, real-time PCR; qPCR, quantitative PCR; IS-PCR, in situ PCR.
Reported prevalence for EBV ranged from 0% to 69% in the indexed literature in our study timeframe, with a median value of 23.5%, as reported in Table 5.
As for the previous viruses, some caveats must be made for the interpretation of the raw prevalence data reported: several studies that used in situ detection techniques actually reported the tumor-infiltrating lymphocytes (TILs) to be the source of the signal, rather than the cancer itself [151]; Khan et al. [151] reported a 48% positivity in their studies, but it was all confined to the tumor-infiltrating lymphocytes. In the study by Aguayo et al. [73], ISH for EBER1 showed that all the breast cancer cases were negative, while Corbex et al. [61] reported that only 1/10 PCR-detected cases showed EBER1 positivity in the cancer nuclei using ISH. Similar results were published by Baltzell et al. [144], that reported a 3% positivity by PCR in their cohort, but failed to confirm any positivity with ISH, and specified also that the PCR positivity had been localized in normal non-cancerous cells. Both Glaser et al. [152] and Hachana et al. [146] reported TILs as the only positive EBV localization and EBER-ISH source, respectively. Mekrazi et al. [129] also reported positivity in TILs rather than tumor cells when analyzing PCR-positive specimens with ISH.
Finally, Peng et al. [63] did report ISH positivity confined to the breast cancer cells, but did not report the number of cases tested with this technique and the percentage in which positivity was detected.
The discrepancy in ISH and IHC rate of positivity has been partially attributed to the quality of RNA material in FFPE specimens, which would contribute to the lower rate of detection by ISH; on this note, Glenn et al. [17] demonstrated a significant presence of EBV in breast cancer when using PCR on fresh frozen DNA extracts; however, when PCR was performed in situ in the same study, the percentage of positivity was actually higher in the control group rather than in the cancer group; the authors do not present an explanation for this finding. In contrast with this finding, Oliveira et al. [27] reported a 7% positivity for EBER1 with ISH, but found a 69% positivity for EBNA1 with immunohistochemistry.
No definitive association with prognostic and predictive factors was found [130,131,138] and the results at our disposal are contrasting: Hachana et al. [146] reported a correlation between EBV DNA presence and negativity for estrogen receptor; however, in the same article, they failed to demonstrate any positivity for EBV with ISH in tumor cells.
Oliveira et al. [27] found a higher prevalence of EBV in low grade and luminal A breast cancer, while both Corbex et al. [61] and Pai et al. [138] reported a higher prevalence of EBV in triple negative breast cancer; similarly, Mashaly et al. [153] reported an association with more aggressive tumors and an increased frequency of EBV subtype D in breast cancer patients; the same subtype was also associated with negativity for progesterone receptor and negativity for HER2. Also Mazouni et al. [148] reported EBV positive breast cancer to be more aggressive with high histological grade and a higher frequency of non-luminal phenotypes, while Zekri et al. [145] did not report any association with histological grade.
Finally, Ballard et al. [141] reported no significant preference for EBV detection in lobular breast cancer against ductal histology, which was instead suggested by Fawzy et al. [150], albeit with very small numbers in the lobular group (8 vs. 32 ductal carcinomas).
Zhang et al. [131] found that EBV positive breast cancer had a high positive expression for PD1/PDL1 and reported worse OS and DFS in EBV-positive patients. On the contrary, Marrão et al. and Joshi et al. [143,149] did not find any significant association between EBV positivity and viral load on OS; paradoxically, Marrão [143] reported that the latent form of EBV infection could confer a survival advantage to the patient. Analogous results were reported by Heng et al. [154] that utilized the Health of Women study database to investigate an association between infectious mononucleosis as a surrogate for EBV infections and breast cancer; the author reported that women diagnosed with infectious mononucleosis before 22 years of age had lower risk of breast cancer (adjusted odds ratio 0.83) than women that never contracted EBV.
El-Naby et al. and Shahi et al. [130,139] reported a higher nodal status in EBV-positive patients. Mostafaei et al. [135] reported higher levels of inflammatory markers such as TNFα in patients with EBV+ breast cancer than in controls.
To complete our review of the relevance of EBV positivity in breast cancer, we analyzed the selected articles to evaluate the difference in prevalence between breast cancer and controls. The data are summarized in Table 6, as follows.
Table 6.
Summary of the prevalence of EBV detection of investigated cases and controls.
| Study | EBV-Positive Cases/Total | EBV-Positive Controls/Total | Statistically Significant? Y/N (p-Value) |
|---|---|---|---|
| Charostad, 2021 [133] | 6/51 (12%) | 1/51 (1%) § | No (p = 0.0599) |
| Golrokh Mofrad, 2020 [134] | 4/59 (7%) | 0/59 (0%) # | No (p = 0.1186) |
| Mostafaei, 2020 [135] | 38/83 (46%) | 5/31 (16%) # | Yes (p = 0.004) |
| Dowran, 2019 [136] | 0/150 (0%) | 0/150 (0%) *,& | // |
| Sharifpour, 2019 [137] | 10/37 (27%) | 4/35 (18%) * | No (p = 0.09) |
| El-Naby, 2017 [139] | 10/42 (24%) | 6/42 (14%) * | No (p = 0.4) |
| Reza, 2015 [106] | 8/100 (8%) with Real time PCR 18/100 (18%) with Eber RNA |
0/100 (0%) § | Yes (p = 0.0068) |
| Richardson, 2015 [142] | 24/70 (34%) | 9/70 (12%) § | Yes (p = 0.0048) |
| Peng, 2014 [63] | 60/100 (60%) | 16/50 (32%) # | Yes (p = 0.0017) |
| Glenn, 2012 [17] | 34/50 (68%) with liquid PCR, 5/27 (19%) with IS-PCR |
14/40 (35%) ^ with liquid PCR, 6/18 (33%) ^ with IS-PCR |
Yes with liquid PCR (p = 0.0028) No with IS-PCR (p = 0.3040) |
| Zekri, 2012 [145] | 32/90 (35%) | 0/20 (0%) ^ | Yes (p = 0.0007) |
| Hachana, 2011 [146] | 33/123 (27%) | 0/123 (0%) § | Yes (p < 0.0001) |
| Kadivar, 2011 [147] | 0/100 (0%) | 0/42 (0%) # | // |
| Lorenzetti, 2010 [127] | 22/71 (31%) | 0/48 (0%) # | Yes (p < 0.0001) |
| Joshi, 2009 [149] | 28/58 (55%) | 0/30 (0%) # | Yes (p < 0.0001) |
| Fawzy, 2008 [150] | 10/40 (25%) | 0/10 (0%) & | No (p = 0.1791) |
* fibroadenomas, # unspecified benign lesions, § normal breast tissue from the same patient, ^ normal breast tissue from healthy patients, & fibrocystic disorder, //, data not available.
Pooling cases and controls together, there is an EBV prevalence of 24% (290/1201) in the cases and of 5% (47/877) in the controls. p-value was significant (p < 0.0001).
Regarding the study design, there was a prevalence of mixed benign lesions (7/16 studies) as control samples. This makes any consideration that goes beyond ‘benign vs. malignant’ very complex, because we cannot make any inference about the contribution of EBV on the pathogenesis of other lesions other than invasive breast carcinoma; for example, the two papers by Sharifpour et al. [137] and El-Naby et al. [139] reported a 18% and 14% rate of positivity also in fibroadenomas.
Also, for EBV, as for the previously reported viruses, the pooled statistical significance of the cases and controls is very strong; however, as for the previously reported viruses, there are important limitations that must be taken into account for interpreting the data. The very frequent report that EBV positivity comes from TILs makes PCR-only studies unreliable.
6. CMV
Cytomegalovirus (CMV) is a frequent virus in humans; around 40 to 90% of the population will contract the infection during their lifetime. The viral cycle consists of alternate lytic and latent phases of infection, where the virus is kept under control by the immune system, and reactivates with inflammatory states or immunodepression; it usually presents with an asymptomatic course or with very mild symptoms in immunocompetent patients, but its reactivation can cause severe and life-threatening disease in immunocompromised hosts, such as pneumonitis, meningitis, colitis, and hepatitis.
The relationship of CMV with cancer is a complex and controversial one, but even if its exact role has not been clarified as of yet in any of the suggested malignancies [155], it has been investigated in relation to various important aspects of carcinogenesis, including tumor microenvironment, epithelial–mesenchymal transition, and overall immune system regulation [156]. It has also been shown to be able to trigger oncogenic transformation in mammary epithelial cells in vitro [157,158] via the development of polyploid giant cells, with downstream activation of the oncogene c-Myc and EZH2 [159,160]. These strains have been referred to as high-risk CMV, referencing the common terminology used for HPV; the same French group was also able to report the isolation of two of these high-risk CMV strains from two triple negative breast cancer biopsies [161]. Kumar et al. [157] reported that infected cells were capable of causing mammary tumors in transfected mice.
Data about CMV reported prevalence in breast cancer is reported in Table 7.
Table 7.
Prevalence of CMV detection in breast cancer samples according to different studies.
| Study | Positive Cases/Total Cases (%) | Technique Used |
|---|---|---|
| Touma, 2023 [162] | 9/109 (8%) for IE protein, 82/108 (76%) for LA protein |
IHC on FFPE TMA |
| Calderon, 2022 [24] | 169/233 (73%) | IHC on FFPE sections |
| Ghaffari, 2021 [132] | 5/72 (7%) | Nested-PCR on FFPE sections |
| Golrokh Mofrad, 2021 [30] | 0/59 (0%) | Nested-PCR on FFPE sections |
| Nakhaie, 2021 [163] | 8/49 (16%) | PCR on CPT |
| Touma, 2021 [164] | 83/111 (75%) for LA protein, 9/111 (8%) for IE protein |
IHC on FFPE sections |
| Costa, 2019 [165] | 49/49 (100%) for IE protein, 11/49 (22%) for LA protein |
PCR on FFPE sections |
| Sepahvand, 2019 [166] | 20/37 (54%) | PCR on FFPE sections |
| El Shazly, 2018 [167] | 11/61 (18%) for CMV DNA 21/61 (34%) for IE protein |
PCR on FFPE sections |
| Richardson, 2015 [142] | 0/70 (0%) | qPCR on CPT |
| Bakhtiyrizadeh, 2017 [42] | 0/150 (0%) | RT-PCR on FFPE sections |
| El-Shinawi, 2013 [168] | 62% | Nested-PCR on FFPE sections |
IHC, immunohistochemistry; FFPE, formalin-fixed, paraffin-embedded; TMA, tissue microarray; CPT, cryopreserved tissue; qPCR, quantitative PCR; RT-PCR, real-time PCR.
Reported prevalence for CMV ranged from 0% to 100% in the indexed literature in our study timeframe, with a median value of 22%, as reported in Table 7.
The heterogeneity of techniques used for virus detection is, as previously mentioned, a strong limit in the reproducibility and correct evaluation of the significance of the reported findings; for example, when confirming IHC results with qPCR, Touma et al. [164] were only able to detect viral DNA in 30% of the tested cases. Therefore, IHC only reports might be considered unreliable. The detection of CMV full genome was reported in some of the works by Herbein’s group [160], but not in other works of the same group, [157], which also makes selection of the PCR primer important and might explain poor detection rates in some of the studies, also depending on the phase in which the virus is detected inside the cell (lytic vs. latent).
El-Shinawi [168] reported a higher frequency of CMV positivity in inflammatory breast cancer than in any other histotype. They also reported an increased activation of NF-κB p65 in CMV-positive inflammatory breast cancer. On a similar note, Costa et al. [165] reported a positive association between extensive positivity for CMV IE protein and the proinflammatory mediators COX-2 and 5-LO.
In their paper, Nakhaie et al. [163] reported not only a significant difference in CMV detection rate between breast cancer tissue and adjacent normal tissue, but also a significant difference in the detection of anti-CMV IgG, which were statistically higher in the breast cancer patients, possibly reinforcing the hypothesis that late exposure to the virus may be linked to the development of breast cancer. However, Pandey et al. [169] reported a contrary result in a larger cohort, in which IgG against CMV glycoprotein-B were higher in the healthy control population than in breast cancer patients.
Touma et al. [162] reported a shorter overall survival (OS) for patients who were positive for CMV IE protein at diagnosis; however, no information about treatment, comorbidity or tumor biology and stage was included in this analysis. Using a regression analysis, only expression of LA protein showed a tendency toward significance for a shorter OS, but that also failed to reach significance.
No association of CMV positivity and lymph node status, tumor dimension or proliferation index was reported [165,167].
To complete our review of the relevance of CMV positivity in breast cancer, we analyzed the selected articles to evaluate the difference in prevalence between breast cancer and controls. The data are summarized in Table 8, as follows.
Table 8.
Summary of the prevalence of CMV detection of investigated cases and controls.
| Study | CMV-Positive Cases/Total | CMV-Positive Controls/Total | Statistically Significant? Y/N (p-Value) |
|---|---|---|---|
| Nakhaie, 2021 [163] | 8/49 (16%) | 1/49 (2%) § | Yes (p = 0.0307) |
| Costa, 2019 [165] | 49/49 (100%) for IE protein, 11/49 (22%) for LA protein |
2/26 (8%) § for IE protein 0/26 (0%) § for LA protein |
Yes (p = 0.0001) for IE protein, (p = 0.0126) for LA protein |
| Sepahvand, 2019 [166] | 20/37 (54%) | 10/35 (26%) * | Yes (p = 0.0340) |
| El Shazly, 2018 [167] | 11/61 (18%) | 1/20 (5%) * | No (p = 0.2765) |
| Bakhtiyrizadeh, 2017 [42] | 0/150 (0%) | 2/150 (1%) # | No (p = 0.4983) |
| Richardson, 2015 [142] | 0/70 (0%) | 2/70 (3%) § | No (p = 0.4964) |
* fibroadenomas, # unspecified benign lesions, § normal breast tissue from the same patient.
Pooling cases and controls together, there is a CMV prevalence of 12% (50/416) in the cases and of 5% (16/350) controls. p-value was significant (p = 0.0003). However, despite this significant statistical result, we feel that the numbers considered are actually too small to draw strong epidemiological inferences.
7. BLV
Bovine leukemia virus (BLV) is a bovine oncogenic retrovirus that causes a disease similar to B cell leukemia. The presence in humans of antibodies that react to BLV antigens supports the theory that a zoonotic transmission of the virus may happen via direct contact or consumption of contaminated products, but a clear transmission mechanism has not been clearly demonstrated, nor has its implication as a causative cancer agent. Moreover, results regarding the actual prevalence of BLV antibodies in serum samples are controversial, ranging from 0% to 74.3% in a recent metanalysis [170]. Some authors believe transmission from cattle to human to be plausible, as the bovine infection is widespread; consumption of raw, unpasteurized milk and raw or undercooked beef might be the vessel, as the virus is rendered non-infectious by heat.
Data about BLV reported prevalence in breast cancer is reported in Table 9.
Table 9.
Prevalence of BLV detection in breast cancer samples according to different studies.
| Study | Positive Cases/Total Cases (%) | Technique Used |
|---|---|---|
| Amato, 2023 [171] | 0/30 (0%) | PCR on CPT |
| Yamanaka, 2022 [172] | 0/23 (0%) | PCR on CPT |
| Adekanmbi, 2021 [173] | 0/238 (0%) | Fluorescence resonance energy transfer-qPCR on blood-derived DNA samples |
| Olaya-Galán, 2021 [174] | 46/75 (61%) | Nested-liquid phase PCR on FFPE sections |
| Delarmelina, 2020 [175] | 47/49 (96%) 46/49 (94%) for the tax gene 28/49 (57%) for the env gene |
Nested-PCR on FFPE |
| Schwingel, 2019 [176] | 22/72 (31%) | PCR on FFPE sections |
| Baltzell, 2018 [36] | 35/61 (57%) | PCR and/or DNA hybridization on FFPE sections |
| Buehring, 2017 [177] | 40/50 (80%) | PCR-ISH on FFPE sections |
| Gillet, 2016 [178] | 0/51 (0%) | Raw DNA sequences from whole genomes of breast tumors and normal breast tissues adjacent to the tumor were retrieved from the NCBI database of genotype and phenotype. |
| Zhang, 2016 [179] | 0/91 (0%) | RT-PCR on unspecified tissue samples |
| Buehring, 2015 [180] | 67/114 (59%) | IS-PCR on FFPE sections |
| Buehring, 2014 [181] | 97/2019 (44%) | IS-PCR on FFPE sections |
CPT, cryopreserved tissue; qPCR, quantitative PCR; FFPE, formalin-fixed, paraffin-embedded; PCR-ISH, PCR-in situ hybridization; RT-PCR, real-time PCR; IS-PCR, in situ PCR.
Reported prevalence for BLV ranged from 0% to 96% in the indexed literature in our study timeframe, with a median value of 37.5%, as reported in Table 9. As previously reported in other sections of this review, a study by Khan et al. [182] was excluded because the case group included benign, malignant and infectious lesions.
As with most of the previously mentioned viruses, we briefly touch again on the topic of PCR limitations, because we feel that it is an important factor to properly analyze the results; in this case, we stress once again the heterogeneous results that primer choice can make; for example, the gene regions gag, pol and env are often eliminated from the DNA as the tumor progresses, while the tax region is conserved longer; this is nicely highlighted by the results by Delarmelina et al. [175], which reported a 94% positivity when using a primer annealing the tax gene, which dropped to 57% when using a primer against the env gene.
Buehring et al. [180] analyzed the prevalence of BLV DNA across a biological gradient of lesions; they included patients with ‘premalignant’ conditions, such as ADH and carcinoma in situ, and reported that 38% of tissues with these lesions were positive for BLV DNA (against 59% of malignant tissues, and 29% of tissues from healthy controls).
Interestingly, Buehring et al. [177] also investigated if a temporal relationship between exposure and cancer development exists; they analyzed 48 subjects, for which a benign breast sample from previous surgery was present. Of these 48 subjects, 28 (58%) developed cancer. Of the patients that developed cancer, 20/28 had BLV DNA detectable in both specimens, and in 3 patients that had a premalignant diagnosis in the first specimen, BLV DNA was detectable both in the premalignant and in the malignant specimen.
No association between BLV DNA detection in breast cancer and predictive and prognostic factors was reported by any of the authors [174,175,176,180].
To complete our review of the relevance of BLV positivity in breast cancer, we analyzed the selected articles to evaluate the difference in prevalence between breast cancer and controls. The data are summarized in Table 10, as follows.
Table 10.
Summary of the prevalence of BLV detection of investigated cases and controls.
| Study | BLV-Positive Cases/Total | BLV-Positive Controls/Total | Statistically Significant? Y/N (p-Value) |
|---|---|---|---|
| Olaya-Galán, 2021 [174] | 46/75 (61%) | 40/83 (48%) # | No (p = 0.1113) |
| Delarmelina, 2020 [175] | 47/49 (96%) 46/49 (94%) for the tax gene 28/49 (57%) for the env gene |
23/39 (59%) ^ 20/39 (51%) ^ for the tax gene 14/39 (36%) ^ for the env gene |
Yes (p < 0.001) |
| Schwingel, 2019 [176] | 22/72 (31%) | 10/72 (14%) ^ | Yes (p = 0.0265) |
| Baltzell, 2018 [36] | 35/61 (57%) | 20/103 (20%) § | Yes (p < 0.0001) |
| Buehring, 2017 [177] | 40/50 (80%) | 19/46 (41%) # | Yes (p = 0.0001) |
| Buehring, 2015 [180] | 67/114 (59%) | 30/104 (29%) ^ | Yes (p < 0.0001) |
# unspecified benign lesions, § normal breast tissue from the same patient, ^ normal breast tissue from healthy patients.
Pooling cases and controls together, there is a BLV prevalence of 57% (238/421) in the cases and of 30% (133/447) controls. p-value was significant (p < 0.0001). However, as for CMV in the previous paragraph, we feel that there are not large enough samples to draw strong epidemiological conclusions.
8. JCV
JCV, or human polyomavirus 2, is a polyomavirus whose asymptomatic infection is detected in 80–90% of immunocompetent adults, and it has been shown to cause progressive multifocal leukoencephalopathy and other disease in immunocompromised patients only. Sequences of the virus have been detected across several neoplasias [183,184,185,186], but the results are controversial and mostly inconclusive.
Two studies investigated the prevalence of JCV in breast cancer.
One study by Dowran et al. [136] failed to detect any virus DNA by PCR in 150 cases and 150 controls. On the other hand, Hachana et al. [187] reported a 23% (28/123) prevalence of JCV in a Tunisian breast cancer cohort, and suggested a correlation to a triple negative phenotype.
The data in the literature do not appear to support a possible contribution of JCV in breast cancer.
9. MCPyV
Merkel cell polyomavirus (MCPyV) is a human polyomavirus, thought to be the causative agent of the majority of cases of Merkel cell carcinoma, a highly aggressive neuroectodermal skin cancer. It appears to be a rather common infection in the general population, that can be detected on healthy skin in 60–90% of various sites, where it infects dermal fibroblasts and persists in a latent, episomal form of the viral genome [188]; infection occurs early in life, through shedding of the virions from healthy skin, either via skin infection or via the respiratory tract.
Data about MCPyV reported prevalence in breast cancer is reported in Table 11.
Table 11.
Prevalence of MCPyV detection in breast cancer samples according to different studies.
| Study | Positive Cases/Total Cases (%) | Technique Used |
|---|---|---|
| Reza, 2015 [106] | 3/100 (3%) | RT-PCR on FFPE sections |
| Corbex, 2014 [61] | 1/123 (1%) | PCR on FFPE sections |
| Peng, 2014 [63] | 14/100 (14%) | Multiplex PCR detected by matrix-assisted laser desorptionionization-time of flight mass spectrometry on CPT |
| Khan, 2012 [189] | 0/58 (0%) | PCR on FFPE sections |
RT-PCR, real-time PCR; FFPE, formalin-fixed, paraffin-embedded; CPT, cryopreserved tissue.
Reported prevalence for MCPyV ranged from 0% to 14% in the indexed literature in our study timeframe, with a median value of 2%, as reported in Table 11.
One study by Peng et al. [63] investigated the prevalence of MCPyV also in healthy controls and reported a positivity rate of 2% (1/50) vs. 14% in neoplastic cases (p value = 0.0210); similar values were also reported by Reza et al. [106] (3% detection rate in cases, 0% detection in the controls).
The data in the literature do not appear to support a possible contribution of MCPyV in breast cancer.
10. Conclusions
To our knowledge, this review represents the most up-to-date work on the topic at hand. While we recognize that it has limitations, such as the heterogeneity of the papers included, that rendered a systematic meta-analysis impossible, and the fact that we only included results from Pubmed, we think that the complete overview that it provides is still of value.
As per our judgment, no evidence appears to exist to suggest that JCV or MCPyV may contribute to the development of breast cancer; for CMV and BLV we feel that, even if some interesting data exist regarding prevalence of infection, case–control studies are too limited to draw strong conclusions.
Regarding the contribution of HPV, MMTV and EBV to breast carcinogenesis, extensive raw data exist in the literature that might support their involvement, but contribution is severely hindered by the small cohorts analyzed, the variety of techniques and samples that are included, and the lack of scientifically rigorous study design. While some evidence might be compelling, and could have an important impact on public health strategies, we advocate for the need of extensive and robust epidemiological studies to advance the knowledge in this field. A more uniform choice of test, with a preference for in situ techniques that allow for a correct visualization of the positivity signal, could improve the confidence in positive results, as well as the design of proper case–control studies on a larger scale; for HPV, we also feel that the introduction of large-scale vaccination campaigns and their possible effect on breast cancer prevention as well should be properly investigated in the next few years.
Author Contributions
Conceptualization, C.R. and M.L.; methodology, C.R.; formal analysis, M.L.; investigation, C.R.; resources, F.S.I., S.C., G.R. and A.L.; writing—original draft preparation, C.R.; writing—review and editing, M.L. and A.L.; supervision, M.L., P.P. and M.P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Rous P. A Sarcoma of the Fowl Transmissible by An Agent Separable from the Tumor Cells. J. Exp. Med. 1911;13:397–411. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bittner J.J. Some Possible Effects of Nursing on the Mammary Gland Tumor Incidence in Mice. Science. 1936;84:162. doi: 10.1126/science.84.2172.162. [DOI] [PubMed] [Google Scholar]
- 3.De Martel C., Georges D., Bray F., Ferlay J., Clifford G.M. Global Burden of Cancer Attributable to Infections in 2018: A Worldwide Incidence Analysis. Lancet Glob. Health. 2020;8:e180–e190. doi: 10.1016/S2214-109X(19)30488-7. [DOI] [PubMed] [Google Scholar]
- 4.Hill A.B. The Environment and Disease: Association or Causation? Proc. R. Soc. Med. 1965;58:295–300. doi: 10.1177/003591576505800503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO Breast Cancer. [(accessed on 25 April 2024)]; Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer.
- 6.Page M.J., McKenzie J.E., Bossuyt P.M., Boutron I., Hoffmann T.C., Mulrow C.D., Shamseer L., Tetzlaff J.M., Akl E.A., Brennan S.E., et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ. 2021;372:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milano G., Guarducci G., Nante N., Montomoli E., Manini I. Human Papillomavirus Epidemiology and Prevention: Is There Still a Gender Gap? Vaccines. 2023;11:1060. doi: 10.3390/vaccines11061060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Checchi M., Mesher D., Panwar K., Anderson A., Beddows S., Soldan K. The Impact of over Ten Years of HPV Vaccination in England: Surveillance of Type-Specific HPV in Young Sexually Active Females. Vaccine. 2023;41:6734–6744. doi: 10.1016/j.vaccine.2023.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Band V., Zajchowski D., Kulesa V., Sager R. Human Papilloma Virus DNAs Immortalize Normal Human Mammary Epithelial Cells and Reduce Their Growth Factor Requirements. Proc. Natl. Acad. Sci. USA. 1990;87:463–467. doi: 10.1073/pnas.87.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akil N., Yasmeen A., Kassab A., Ghabreau L., Darnel A.D., Al Moustafa A.-E. High-Risk Human Papillomavirus Infections in Breast Cancer in Syrian Women and Their Association with Id-1 Expression: A Tissue Microarray Study. Br. J. Cancer. 2008;99:404–407. doi: 10.1038/sj.bjc.6604503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Islam M.S., Chakraborty B., Panda C.K. Human Papilloma Virus (HPV) Profiles in Breast Cancer: Future Management. Ann. Transl. Med. 2020;8:650. doi: 10.21037/atm-19-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreira D.A., Tayyar Y., Idris A., McMillan N.A.J. A “Hit-and-Run” Affair—A Possible Link for Cancer Progression in Virally Driven Cancers. Biochim. Biophys. Acta (BBA) Rev. Cancer. 2021;1875:188476. doi: 10.1016/j.bbcan.2020.188476. [DOI] [PubMed] [Google Scholar]
- 13.Lasagna A., Cassaniti I., Sacchi P., Figini S., Baldanti F., Bruno R., Pedrazzoli P. The ‘Hit-and-Run’ Strategy and Viral Carcinogenesis. Future Oncol. 2023;19:341–344. doi: 10.2217/fon-2022-1171. [DOI] [PubMed] [Google Scholar]
- 14.Glenn W.K., Whitaker N.J., Lawson J.S. High Risk Human Papillomavirus and Epstein Barr Virus in Human Breast Milk. BMC Res. Notes. 2012;5:477. doi: 10.1186/1756-0500-5-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cazzaniga M., Gheit T., Casadio C., Khan N., Macis D., Valenti F., Miller M.J., Sylla B.S., Akiba S., Bonanni B., et al. Analysis of the Presence of Cutaneous and Mucosal Papillomavirus Types in Ductal Lavage Fluid, Milk and Colostrum to Evaluate Its Role in Breast Carcinogenesis. Breast Cancer Res. Treat. 2009;114:599–605. doi: 10.1007/s10549-008-0040-3. [DOI] [PubMed] [Google Scholar]
- 16.Heng B., Glenn W.K., Ye Y., Tran B., Delprado W., Lutze-Mann L., Whitaker N.J., Lawson J.S. Human Papilloma Virus Is Associated with Breast Cancer. Br. J. Cancer. 2009;101:1345–1350. doi: 10.1038/sj.bjc.6605282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glenn W.K., Heng B., Delprado W., Iacopetta B., Whitaker N.J., Lawson J.S. Epstein-Barr Virus, Human Papillomavirus and Mouse Mammary Tumour Virus as Multiple Viruses in Breast Cancer. PLoS ONE. 2012;7:e48788. doi: 10.1371/journal.pone.0048788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preti M., Rosso S., Micheletti L., Libero C., Sobrato I., Giordano L., Busso P., Gallio N., Cosma S., Bevilacqua F., et al. Risk of HPV-Related Extra-Cervical Cancers in Women Treated for Cervical Intraepithelial Neoplasia. BMC Cancer. 2020;20:972. doi: 10.1186/s12885-020-07452-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mareti E., Chatzakis C., Pratilas G.-C., Liberis A., Vavoulidis E., Papanastasiou A., Dampali R., Daniilidis A., Zepiridis L., Dinas K. Human Papillomavirus in Breast Cancer of Patients with Cervical Intraepithelial Neoplasia or Cervical Cancer History. A Syst. Rev. Meta Analysis. J. BUON. 2021;26:707–713. [PubMed] [Google Scholar]
- 20.Frega A., Lorenzon L., Bononi M., De Cesare A., Ciardi A., Lombardi D., Assorgi C., Gentile M., Moscarini M., Torrisi M.R., et al. Evaluation of E6 and E7 mRNA Expression in HPV DNA Positive Breast Cancer. Eur. J. Gynaecol. Oncol. 2012;33:164–167. [PubMed] [Google Scholar]
- 21.Haghighi Z.M.S., Tabatabaei T., Rafigh M., Karampour R., Babaei F., Amjad Z.S., Payandeh M., Roozgari M., Bayat M., Doroudian M., et al. Human Papillomavirus Maybe Is a Critical Player in the Regulation of Chemoresistance Related Factors (P53, Rb, TWIST, Bcl-2, Bcl-XL, c-IAP2, Cytochrome C, and Caspase 3) in Breast Cancer. Pathol. Res. Pract. 2023;248:154653. doi: 10.1016/j.prp.2023.154653. [DOI] [PubMed] [Google Scholar]
- 22.Maroga N., Mokoena T., Ledibane N., Musekiwa A., Bida M., Kgomo M., Lebelo R. Profile of Human Papillomavirus Genotypes in Breast and Oesophageal Cancer Patients in Pretoria, South Africa. S. Afr. Med. J. 2023;113:49–54. doi: 10.7196/SAMJ.2023.v113i7.560. [DOI] [PubMed] [Google Scholar]
- 23.Alinezhadi M., Makvandi M., Kaydani G.A., Jazayeri S.N., Charostad J., Talaeizadeh A.T., Angali K.A. Detection of High-Risk Human Papillomavirus DNA in Invasive Ductal Carcinoma Specimens. Asian Pac. J. Cancer Prev. 2022;23:3201–3207. doi: 10.31557/APJCP.2022.23.9.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calderon G., Castaneda C.A., Castillo M., Sanchez J., Bernabe L., Suarez N., Tello K., Torres E., Cotrina J.M., Dunstan J., et al. Human Papillomavirus, Cytomegalovirus Infection and P16 Staining in Breast Tumors from Peruvian Women. Asian Pac. J. Cancer Prev. 2022;23:1571–1576. doi: 10.31557/APJCP.2022.23.5.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta I., Al-Sarraf R., Farghaly H., Vranic S., Sultan A.A., Al-Thawadi H., Al Moustafa A.-E., Al-Farsi H.F. Incidence of HPVs, EBV, and MMTV-Like Virus in Breast Cancer in Qatar. Intervirology. 2022;65:188–194. doi: 10.1159/000525277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mamani-Zapana E., Vilcapoma-Diaz M., Ortiz-Muchotrigo N. Detection of human papilloma virus in breast cancer biopsies using PCR and immunohistochemistry at the santa rosa hospital during 2019. Rev. Peru. Med. Exp. Salud Publica. 2022;39:450–455. doi: 10.17843/rpmesp.2022.394.11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliveira E.S.D., Ferreira M.V.P., Rahal P., Castelo Branco M.B., Rabenhorst S.H.B. High Frequency of Epstein-Barr Virus and Absence of Papillomavirus in Breast Cancer Patients from Brazilian Northeast. Asian Pac. J. Cancer Prev. 2022;23:2351–2359. doi: 10.31557/APJCP.2022.23.7.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charostad J., Azaran A., Nakhaei M., Astani A., Kaydani G.A., Motamedfar A., Makvandi M. Upregulation of Interleukin-6 in HPV-Positive Breast Cancer Patients. Iran. J. Immunol. 2021;18:315–330. doi: 10.22034/IJI.2021.89107.1930. [DOI] [PubMed] [Google Scholar]
- 29.Elagali A.M., Suliman A.A., Altayeb M., Dannoun A.I., Parine N.R., Sakr H.I., Suliman H.S., Motawee M.E. Human Papillomavirus, Gene Mutation and Estrogen and Progesterone Receptors in Breast Cancer: A Cross-Sectional Study. Pan Afr. Med. J. 2021;38:43. doi: 10.11604/pamj.2021.38.43.22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Golrokh Mofrad M., Sadigh Z.A., Ainechi S., Faghihloo E. Detection of Human Papillomavirus Genotypes, Herpes Simplex, Varicella Zoster and Cytomegalovirus in Breast Cancer Patients. Virol. J. 2021;18:25. doi: 10.1186/s12985-021-01498-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta I., Ulamec M., Peric-Balja M., Ramic S., Al Moustafa A.-E., Vranic S., Al-Farsi H.F. Presence of High-Risk HPVs, EBV, and MMTV in Human Triple-Negative Breast Cancer. Hum. Vaccin. Immunother. 2021;17:4457–4466. doi: 10.1080/21645515.2021.1975452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H., Idrovo J.P., Cao J., Roychoudhury S., Navale P., Auguste L.J., Bhuiya T., Sheikh-Fayyaz S. Human Papillomavirus (HPV) Detection by Chromogenic In Situ Hybridization (CISH) and P16 Immunohistochemistry (IHC) in Breast Intraductal Papilloma and Breast Carcinoma. Clin. Breast Cancer. 2021;21:e638–e646. doi: 10.1016/j.clbc.2021.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Sher G., Salman N.A., Kulinski M., Fadel R.A., Gupta V.K., Anand A., Gehani S., Abayazeed S., Al-Yahri O., Shahid F., et al. Prevalence and Type Distribution of High-Risk Human Papillomavirus (HPV) in Breast Cancer: A Qatar Based Study. Cancers. 2020;12:1528. doi: 10.3390/cancers12061528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tawfeik A.M., Mora A., Osman A., Moneer M.M., El-Sheikh N., Elrefaei M. Frequency of CD4+ Regulatory T Cells, CD8+ T Cells, and Human Papilloma Virus Infection in Egyptian Women with Breast Cancer. Int. J. Immunopathol. Pharmacol. 2020;34:2058738420966822. doi: 10.1177/2058738420966822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balci F.L., Uras C., Feldman S.M. Is Human Papillomavirus Associated with Breast Cancer or Papilloma Presenting with Pathologic Nipple Discharge? Cancer Treat. Res. Commun. 2019;19:100122. doi: 10.1016/j.ctarc.2019.100122. [DOI] [PubMed] [Google Scholar]
- 36.Baltzell K.A., Shen H.M., Krishnamurthy S., Sison J.D., Nuovo G.J., Buehring G.C. Bovine Leukemia Virus Linked to Breast Cancer but Not Coinfection with Human Papillomavirus: Case-Control Study of Women in Texas. Cancer. 2018;124:1342–1349. doi: 10.1002/cncr.31169. [DOI] [PubMed] [Google Scholar]
- 37.Cavalcante J.R., Pinheiro L.G.P., de Almeida P.R.C., Ferreira M.V.P., Cruz G.A., Campelo T.A., Silva C.S., Lima L.N.G.C., de Oliveira B.M.K., Lima L.M., et al. Association of Breast Cancer with Human Papillomavirus (HPV) Infection in Northeast Brazil: Molecular Evidence. Clinics. 2018;73:e465. doi: 10.6061/clinics/2018/e465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghaffari H., Nafissi N., Hashemi-Bahremani M., Alebouyeh M.R., Tavakoli A., Javanmard D., Bokharaei-Salim F., Mortazavi H.S., Monavari S.H. Molecular Prevalence of Human Papillomavirus Infection among Iranian Women with Breast Cancer. Breast Dis. 2018;37:207–213. doi: 10.3233/BD-180333. [DOI] [PubMed] [Google Scholar]
- 39.Habyarimana T., Attaleb M., Mazarati J.B., Bakri Y., El Mzibri M. Detection of Human Papillomavirus DNA in Tumors from Rwandese Breast Cancer Patients. Breast Cancer. 2018;25:127–133. doi: 10.1007/s12282-018-0831-2. [DOI] [PubMed] [Google Scholar]
- 40.Kouloura A., Nicolaidou E., Misitzis I., Panotopoulou E., Kassiani T., Smyrniotis V., Corso G., Veronesi P., Arkadopoulos N. HPV Infection and Breast Cancer. Results of a Microarray Approach. Breast. 2018;40:165–169. doi: 10.1016/j.breast.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 41.Malekpour Afshar R., Balar N., Mollaei H.R., Arabzadeh S.A., Iranpour M. Low Prevalence of Human Papilloma Virus in Patients with Breast Cancer, Kerman; Iran. Asian Pac. J. Cancer Prev. 2018;19:3039–3044. doi: 10.31557/APJCP.2018.19.11.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bakhtiyrizadeh S., Hosseini S.Y., Yaghobi R., Safaei A., Sarvari J. Almost Complete Lack of Human Cytomegalovirus and Human Papillomaviruses Genome in Benign and Malignant Breast Lesions in Shiraz, Southwest of Iran. Asian Pac. J. Cancer Prev. 2017;18:3319–3324. doi: 10.22034/APJCP.2017.18.12.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delgado-García S., Martínez-Escoriza J.-C., Alba A., Martín-Bayón T.-A., Ballester-Galiana H., Peiró G., Caballero P., Ponce-Lorenzo J. Presence of Human Papillomavirus DNA in Breast Cancer: A Spanish Case-Control Study. BMC Cancer. 2017;17:320. doi: 10.1186/s12885-017-3308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Islam S., Dasgupta H., Roychowdhury A., Bhattacharya R., Mukherjee N., Roy A., Mandal G.K., Alam N., Biswas J., Mandal S., et al. Study of Association and Molecular Analysis of Human Papillomavirus in Breast Cancer of Indian Patients: Clinical and Prognostic Implication. PLoS ONE. 2017;12:e0172760. doi: 10.1371/journal.pone.0172760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Naushad W., Surriya O., Sadia H. Prevalence of EBV, HPV and MMTV in Pakistani Breast Cancer Patients: A Possible Etiological Role of Viruses in Breast Cancer. Infect. Genet. Evol. 2017;54:230–237. doi: 10.1016/j.meegid.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 46.Salman N.A., Davies G., Majidy F., Shakir F., Akinrinade H., Perumal D., Ashrafi G.H. Association of High Risk Human Papillomavirus and Breast Cancer: A UK Based Study. Sci. Rep. 2017;7:43591. doi: 10.1038/srep43591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y.-W., Zhang K., Zhao S., Lv Y., Zhu J., Liu H., Feng J., Liang W., Ma R., Wang J. HPV Status and Its Correlation with BCL2, P21, P53, Rb, and Survivin Expression in Breast Cancer in a Chinese Population. BioMed Res. Int. 2017;2017:6315392. doi: 10.1155/2017/6315392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y.X., Li Y.Z., Zhang Z.Y., Wang J.Q., Cui J., Qian X.L. HPV16 E6 Promotes Breast Cancer Proliferation via Upregulation of COX-2 Expression. BioMed Res. Int. 2017;2017:2948467. doi: 10.1155/2017/2948467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doosti M., Bakhshesh M., Zahir S.T., Shayestehpour M., Karimi-Zarchi M. Lack of Evidence for a Relationship between High Risk Human Papillomaviruses and Breast Cancer in Iranian Patients. Asian Pac. J. Cancer Prev. 2016;17:4357–4361. [PubMed] [Google Scholar]
- 50.Ilahi N.E., Anwar S., Noreen M., Hashmi S.N., Murad S. Detection of Human Papillomavirus-16 DNA in Archived Clinical Samples of Breast and Lung Cancer Patients from North Pakistan. J. Cancer Res. Clin. Oncol. 2016;142:2497–2502. doi: 10.1007/s00432-016-2251-z. [DOI] [PubMed] [Google Scholar]
- 51.Yan C., Teng Z.P., Chen Y.X., Shen D.H., Li J.T., Zeng Y. Viral Etiology Relationship between Human Papillomavirus and Human Breast Cancer and Target of Gene Therapy. Biomed. Environ. Sci. 2016;29:331–339. doi: 10.3967/bes2016.043. [DOI] [PubMed] [Google Scholar]
- 52.Wang D., Fu L., Shah W., Zhang J., Yan Y., Ge X., He J., Wang Y., Li X. Presence of High Risk HPV DNA but Indolent Transcription of E6/E7 Oncogenes in Invasive Ductal Carcinoma of Breast. Pathol. Res. Pract. 2016;212:1151–1156. doi: 10.1016/j.prp.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 53.Li J., Ding J., Zhai K. Detection of Human Papillomavirus DNA in Patients with Breast Tumor in China. PLoS ONE. 2015;10:e0136050. doi: 10.1371/journal.pone.0136050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fimereli D., Gacquer D., Fumagalli D., Salgado R., Rothé F., Larsimont D., Sotiriou C., Detours V. No Significant Viral Transcription Detected in Whole Breast Cancer Transcriptomes. BMC Cancer. 2015;15:147. doi: 10.1186/s12885-015-1176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu L., Wang D., Shah W., Wang Y., Zhang G., He J. Association of Human Papillomavirus Type 58 with Breast Cancer in Shaanxi Province of China. J. Med. Virol. 2015;87:1034–1040. doi: 10.1002/jmv.24142. [DOI] [PubMed] [Google Scholar]
- 56.Gannon O.M., Antonsson A., Milevskiy M., Brown M.A., Saunders N.A., Bennett I.C. No Association between HPV Positive Breast Cancer and Expression of Human Papilloma Viral Transcripts. Sci. Rep. 2015;5:18081. doi: 10.1038/srep18081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Salehpour M., Tayyebi Meibodi N., Teimourpour R., Ghorani-Azam A., Sepahi S., Rostami S., Meshkat Z. Frequency of Human Papillomavirus Genotypes 6, 11, 16, 18 And 31 in Paraffin-Embedded Tissue Samples of Invasive Breast Carcinoma, North- East of Iran. Iran. J. Pathol. 2015;10:192–198. [PMC free article] [PubMed] [Google Scholar]
- 58.Vernet-Tomas M., Mena M., Alemany L., Bravo I., De Sanjosé S., Nicolau P., Bergueiro A., Corominas J.M., Serrano S., Carreras R., et al. Human Papillomavirus and Breast Cancer: No Evidence of Association in a Spanish Set of Cases. Anticancer Res. 2015;35:851–856. [PubMed] [Google Scholar]
- 59.Ahangar-Oskouee M., Shahmahmoodi S., Jalilvand S., Mahmoodi M., Ziaee A.A., Esmaeili H.-A., Keshtvarz M., Pishraft-Sabet L., Yousefi M., Mollaei-Kandelous Y., et al. No Detection of “high-Risk” Human Papillomaviruses in a Group of Iranian Women with Breast Cancer. Asian Pac. J. Cancer Prev. 2014;15:4061–4065. doi: 10.7314/apjcp.2014.15.9.4061. [DOI] [PubMed] [Google Scholar]
- 60.Ali S.H.M., Al-Alwan N., Al-Alwany S.H.M. Detection and Genotyping of Human Papillomavirus in Breast Cancer Tissues from Iraqi Patients. East. Mediterr. Health J. 2014;20:372–377. doi: 10.26719/2014.20.6.372. [DOI] [PubMed] [Google Scholar]
- 61.Corbex M., Bouzbid S., Traverse-Glehen A., Aouras H., McKay-Chopin S., Carreira C., Lankar A., Tommasino M., Gheit T. Prevalence of Papillomaviruses, Polyomaviruses, and Herpesviruses in Triple-Negative and Inflammatory Breast Tumors from Algeria Compared with Other Types of Breast Cancer Tumors. PLoS ONE. 2014;9:e114559. doi: 10.1371/journal.pone.0114559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manzouri L., Salehi R., Shariatpanahi S., Rezaie P. Prevalence of Human Papilloma Virus among Women with Breast Cancer since 2005-2009 in Isfahan. Adv. Biomed. Res. 2014;3:75. doi: 10.4103/2277-9175.125873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Peng J., Wang T., Zhu H., Guo J., Li K., Yao Q., Lv Y., Zhang J., He C., Chen J., et al. Multiplex PCR/Mass Spectrometry Screening of Biological Carcinogenic Agents in Human Mammary Tumors. J. Clin. Virol. 2014;61:255–259. doi: 10.1016/j.jcv.2014.07.010. [DOI] [PubMed] [Google Scholar]
- 64.Piana A.F., Sotgiu G., Muroni M.R., Cossu-Rocca P., Castiglia P., De Miglio M.R. HPV Infection and Triple-Negative Breast Cancers: An Italian Case-Control Study. Virol. J. 2014;11:190. doi: 10.1186/s12985-014-0190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Herrera-Goepfert R., Vela-Chávez T., Carrillo-García A., Lizano-Soberón M., Amador-Molina A., Oñate-Ocaña L.F., Hallmann R.S.-R. High-Risk Human Papillomavirus (HPV) DNA Sequences in Metaplastic Breast Carcinomas of Mexican Women. BMC Cancer. 2013;13:445. doi: 10.1186/1471-2407-13-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hossein R., Behzad S., Tahar M., Azadeh N.A. Prevalence of Human Papillomavirus Genotypes Associated with Cervical and Breast Cancers in Iran. Monoclon. Antibodies Immunodiagn. Immunother. 2013;32:399–403. doi: 10.1089/mab.2013.0047. [DOI] [PubMed] [Google Scholar]
- 67.Liang W., Wang J., Wang C., Lv Y., Gao H., Zhang K., Liu H., Feng J., Wang L., Ma R. Detection of High-Risk Human Papillomaviruses in Fresh Breast Cancer Samples Using the Hybrid Capture 2 Assay. J. Med. Virol. 2013;85:2087–2092. doi: 10.1002/jmv.23703. [DOI] [PubMed] [Google Scholar]
- 68.Pereira Suarez A.L., Lorenzetti M.A., Gonzalez Lucano R., Cohen M., Gass H., Martinez Vazquez P., Gonzalez P., Preciado M.V., Chabay P. Presence of Human Papilloma Virus in a Series of Breast Carcinoma from Argentina. PLoS ONE. 2013;8:e61613. doi: 10.1371/journal.pone.0061613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baltzell K., Buehring G.C., Krishnamurthy S., Kuerer H., Shen H.M., Sison J.D. Limited evidence of human papillomavirus on breast tissue using molecular in situ methods. Cancer. 2012;118:1212–1220. doi: 10.1002/cncr.26389. Erratum in Cancer 2012, 118, 2561. [DOI] [PubMed] [Google Scholar]
- 70.Chang P., Wang T., Yao Q., Lv Y., Zhang J., Guo W., Wang L., Chen J. Absence of Human Papillomavirus in Patients with Breast Cancer in North-West China. Med. Oncol. 2012;29:521–525. doi: 10.1007/s12032-011-9945-5. [DOI] [PubMed] [Google Scholar]
- 71.Herrera-Romano L., Fernández-Tamayo N., Gómez-Conde E., Reyes-Cardoso J.M., Ortiz-Gutierrez F., Ceballos G., Valdivia A., Piña P., Salcedo M. Absence of Human Papillomavirus Sequences in Epithelial Breast Cancer in a Mexican Female Population. Med. Oncol. 2012;29:1515–1517. doi: 10.1007/s12032-011-0059-x. [DOI] [PubMed] [Google Scholar]
- 72.Sigaroodi A., Nadji S.A., Naghshvar F., Nategh R., Emami H., Velayati A.A. Human Papillomavirus Is Associated with Breast Cancer in the North Part of Iran. Sci. World J. 2012;2012:837191. doi: 10.1100/2012/837191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aguayo F., Khan N., Koriyama C., González C., Ampuero S., Padilla O., Solís L., Eizuru Y., Corvalán A., Akiba S. Human Papillomavirus and Epstein-Barr Virus Infections in Breast Cancer from Chile. Infect. Agents Cancer. 2011;6:7. doi: 10.1186/1750-9378-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Antonsson A., Spurr T.P., Chen A.C., Francis G.D., McMillan N.A.J., Saunders N.A., Law M., Bennett I.C. High Prevalence of Human Papillomaviruses in Fresh Frozen Breast Cancer Samples. J. Med. Virol. 2011;83:2157–2163. doi: 10.1002/jmv.22223. [DOI] [PubMed] [Google Scholar]
- 75.Ghaffari S.R., Sabokbar T., Meshkat Z., Fereidooni F., Dastan J., Rafati M., Zendehdel K. Tracing Human Papilloma Virus in Breast Tumors of Iranian Breast Cancer Patients. Breast J. 2011;17:218–219. doi: 10.1111/j.1524-4741.2010.01053.x. [DOI] [PubMed] [Google Scholar]
- 76.Hedau S., Kumar U., Hussain S., Shukla S., Pande S., Jain N., Tyagi A., Deshpande T., Bhat D., Mir M.M., et al. Breast Cancer and Human Papillomavirus Infection: No Evidence of HPV Etiology of Breast Cancer in Indian Women. BMC Cancer. 2011;11:27. doi: 10.1186/1471-2407-11-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mou X., Chen L., Liu F., Shen Y., Wang H., Li Y., Yuan L., Lin J., Lin J., Teng L., et al. Low Prevalence of Human Papillomavirus (HPV) in Chinese Patients with Breast Cancer. J. Int. Med. Res. 2011;39:1636–1644. doi: 10.1177/147323001103900506. [DOI] [PubMed] [Google Scholar]
- 78.Silva R.G., da Silva B.B. No Evidence for an Association of Human Papillomavirus and Breast Carcinoma. Breast Cancer Res. Treat. 2011;125:261–264. doi: 10.1007/s10549-010-1129-z. [DOI] [PubMed] [Google Scholar]
- 79.Hachana M., Ziadi S., Amara K., Toumi I., Korbi S., Trimeche M. No Evidence of Human Papillomavirus DNA in Breast Carcinoma in Tunisian Patients. Breast. 2010;19:541–544. doi: 10.1016/j.breast.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 80.Yavuzer D., Salepci T., Karadayi N., Baloglu H., Kucukodaci Z. Human Papillomavirus Is Not Associated with Breast Carcinoma. Breast Cancer Res. Treat. 2010;122:899–900. doi: 10.1007/s10549-010-0963-3. [DOI] [PubMed] [Google Scholar]
- 81.de León D.C., Montiel D.P., Nemcova J., Mykyskova I., Turcios E., Villavicencio V., Cetina L., Coronel A., Hes O. Human Papillomavirus (HPV) in Breast Tumors: Prevalence in a Group of Mexican Patients. BMC Cancer. 2009;9:26. doi: 10.1186/1471-2407-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He Q., Zhang S.-Q., Chu Y.-L., Jia X.-L., Wang X.-L. The Correlations between HPV16 Infection and Expressions of C-erbB-2 and Bcl-2 in Breast Carcinoma. Mol. Biol. Rep. 2009;36:807–812. doi: 10.1007/s11033-008-9249-9. [DOI] [PubMed] [Google Scholar]
- 83.Mendizabal-Ruiz A.P., Morales J.A., Ramírez-Jirano L.J., Padilla-Rosas M., Morán-Moguel M.C., Montoya-Fuentes H. Low Frequency of Human Papillomavirus DNA in Breast Cancer Tissue. Breast Cancer Res. Treat. 2009;114:189–194. doi: 10.1007/s10549-008-9989-1. [DOI] [PubMed] [Google Scholar]
- 84.de Cremoux P., Thioux M., Lebigot I., Sigal-Zafrani B., Salmon R., Sastre-Garau X., Institut Curie Breast Group No Evidence of Human Papillomavirus DNA Sequences in Invasive Breast Carcinoma. Breast Cancer Res. Treat. 2008;109:55–58. doi: 10.1007/s10549-007-9626-4. [DOI] [PubMed] [Google Scholar]
- 85.Duò D., Ghimenti C., Migliora P., Pavanelli M.C., Mastracci L., Angeli G. Identification and Characterization of Human Papillomavirus DNA Sequences in Italian Breast Cancer Patients by PCR and Line Probe Assay Reverse Hybridization. Mol. Med. Rep. 2008;1:673–677. doi: 10.3892/mmr_00000011. [DOI] [PubMed] [Google Scholar]
- 86.Khan N.A., Castillo A., Koriyama C., Kijima Y., Umekita Y., Ohi Y., Higashi M., Sagara Y., Yoshinaka H., Tsuji T., et al. Human Papillomavirus Detected in Female Breast Carcinomas in Japan. Br. J. Cancer. 2008;99:408–414. doi: 10.1038/sj.bjc.6604502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ren C., Zeng K., Wu C., Mu L., Huang J., Wang M. Human Papillomavirus Infection Increases the Risk of Breast Carcinoma: A Large-Scale Systemic Review and Meta-Analysis of Case-Control Studies. Gland. Surg. 2019;8:486–500. doi: 10.21037/gs.2019.09.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Karachalios C., Petousis S., Margioula-Siarkou C., Dinas K. Human Papillomaviruses and Breast Cancer: A Systematic Review and Meta-analysis. Oncol. Lett. 2023;27:75. doi: 10.3892/ol.2023.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Malhone C., Longatto-Filho A., Filassi J.R. Is Human Papilloma Virus Associated with Breast Cancer? A Review of the Molecular Evidence. Acta Cytol. 2018;62:166–177. doi: 10.1159/000487700. [DOI] [PubMed] [Google Scholar]
- 90.Gumus M., Yumuk P.F., Salepci T., Aliustaoglu M., Dane F., Ekenel M., Basaran G., Kaya H., Barisik N., Turhal N.S. HPV DNA Frequency and Subset Analysis in Human Breast Cancer Patients’ Normal and Tumoral Tissue Samples. J. Exp. Clin. Cancer Res. 2006;25:515–521. [PubMed] [Google Scholar]
- 91.Kulka J., Kovalszky I., Svastics E., Berta M., Füle T. Lymphoepithelioma-like Carcinoma of the Breast: Not Epstein-Barr Virus-, but Human Papilloma Virus-Positive. Hum. Pathol. 2008;39:298–301. doi: 10.1016/j.humpath.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 92.Melana S.M., Nepomnaschy I., Hasa J., Djougarian A., Djougarian A., Holland J.F., Pogo B.G.T. Detection of Human Mammary Tumor Virus Proteins in Human Breast Cancer Cells. J. Virol. Methods. 2010;163:157–161. doi: 10.1016/j.jviromet.2009.09.015. [DOI] [PubMed] [Google Scholar]
- 93.May F.E., Westley B.R., Rochefort H., Buetti E., Diggelmann H. Mouse Mammary Tumour Virus Related Sequences Are Present in Human DNA. Nucleic Acids Res. 1983;11:4127–4139. doi: 10.1093/nar/11.12.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Indik S., Günzburg W.H., Kulich P., Salmons B., Rouault F. Rapid Spread of Mouse Mammary Tumor Virus in Cultured Human Breast Cells. Retrovirology. 2007;4:73. doi: 10.1186/1742-4690-4-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stewart T.H., Sage R.D., Stewart A.F., Cameron D.W. Breast Cancer Incidence Highest in the Range of One Species of House Mouse, Mus Domesticus. Br. J. Cancer. 2000;82:446–451. doi: 10.1054/bjoc.1999.0941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mazzanti C.M., Lessi F., Armogida I., Zavaglia K., Franceschi S., Al Hamad M., Roncella M., Ghilli M., Boldrini A., Aretini P., et al. Human Saliva as Route of Inter-Human Infection for Mouse Mammary Tumor Virus. Oncotarget. 2015;6:18355–18363. doi: 10.18632/oncotarget.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Johal H., Ford C., Glenn W., Heads J., Lawson J., Rawlinson W. Mouse Mammary Tumor like Virus Sequences in Breast Milk from Healthy Lactating Women. Breast Cancer Res. Treat. 2011;129:149–155. doi: 10.1007/s10549-011-1421-6. [DOI] [PubMed] [Google Scholar]
- 98.Fekete Z., Tertan B.O., Raduly L., Eniu D.T., Buiga R., Galatar M., Berindan-Neagoe I. Prevalence of MMTV-like Sequences in Breast Cancer Samples in Romanian Patients-There Is a Geographic Difference Compared to the Western World. Infect. Agent. Cancer. 2023;18:39. doi: 10.1186/s13027-023-00486-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Khalid H.F., Ali A., Fawad N., Rafique S., Ullah I., Rehman G., Shams M.U., Idrees M. MMTV-LIKE Virus and c-Myc over-Expression Are Associated with Invasive Breast Cancer. Infect. Genet. Evol. 2021;91:104827. doi: 10.1016/j.meegid.2021.104827. [DOI] [PubMed] [Google Scholar]
- 100.de Sousa Pereira N., Akelinghton Freire Vitiello G., Karina Banin-Hirata B., Scantamburlo Alves Fernandes G., José Sparça Salles M., Karine Amarante M., Angelica Ehara Watanabe M. Mouse Mammary Tumor Virus (MMTV)-Like Env Sequence in Brazilian Breast Cancer Samples: Implications in Clinicopathological Parameters in Molecular Subtypes. Int. J. Environ. Res. Public Health. 2020;17:9496. doi: 10.3390/ijerph17249496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Naccarato A.G., Lessi F., Zavaglia K., Scatena C., Al Hamad M.A., Aretini P., Menicagli M., Roncella M., Ghilli M., Caligo M.A., et al. Mouse Mammary Tumor Virus (MMTV)—Like Exogenous Sequences Are Associated with Sporadic but Not Hereditary Human Breast Carcinoma. Aging. 2019;11:7236–7241. doi: 10.18632/aging.102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seo I., Cho J.-H., Lee M.-H., Park W.-J., Kwon S.-Y., Lee J.-H. Clinical and Prognostic Value of Human Mammary Tumor Virus in Korean Patients with Breast Carcinoma. Ann. Clin. Lab. Sci. 2019;49:171–174. [PubMed] [Google Scholar]
- 103.Al Dossary R., Alkharsah K.R., Kussaibi H. Prevalence of Mouse Mammary Tumor Virus (MMTV)-like Sequences in Human Breast Cancer Tissues and Adjacent Normal Breast Tissues in Saudi Arabia. BMC Cancer. 2018;18:170. doi: 10.1186/s12885-018-4074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Motamedifar M., Saki M., Ghaderi A. Lack of Association of Mouse Mammary Tumor Virus-like Sequences in Iranian Breast Cancer Patients. Med. Princ. Pract. 2012;21:244–248. doi: 10.1159/000334572. [DOI] [PubMed] [Google Scholar]
- 105.Shariatpanahi S., Farahani N., Salehi A.R., Salehi R. High Prevalence of Mouse Mammary Tumor Virus-like Gene Sequences in Breast Cancer Samples of Iranian Women. Nucleosides Nucleotides Nucleic Acids. 2017;36:621–630. doi: 10.1080/15257770.2017.1360498. [DOI] [PubMed] [Google Scholar]
- 106.Reza M.A., Reza M.H., Mahdiyeh L., Mehdi F., Hamid Z.N. Evaluation Frequency of Merkel Cell Polyoma, Epstein-Barr and Mouse Mammary Tumor Viruses in Patients with Breast Cancer in Kerman, Southeast of Iran. Asian Pac. J. Cancer Prev. 2015;16:7351–7357. doi: 10.7314/apjcp.2015.16.16.7351. [DOI] [PubMed] [Google Scholar]
- 107.Ahangar Oskouee M., Shahmahmoodi S., Jalilvand S., Mahmoodi M., Ziaee A.-A., Esmaeili H.A., Mokhtari-Azad T., Yousefi M., Mollaei-Kandelous Y., Nategh R. No Evidence of Mammary Tumor Virus Env Gene-like Sequences among Iranian Women with Breast Cancer. Intervirology. 2014;57:353–356. doi: 10.1159/000366280. [DOI] [PubMed] [Google Scholar]
- 108.Naushad W., Bin Rahat T., Gomez M.K., Ashiq M.T., Younas M., Sadia H. Detection and Identification of Mouse Mammary Tumor Virus-like DNA Sequences in Blood and Breast Tissues of Breast Cancer Patients. Tumor Biol. 2014;35:8077–8086. doi: 10.1007/s13277-014-1972-3. [DOI] [PubMed] [Google Scholar]
- 109.Morales-Sánchez A., Molina-Muñoz T., Martínez-López J.L.E., Hernández-Sancén P., Mantilla A., Leal Y.A., Torres J., Fuentes-Pananá E.M. No Association between Epstein-Barr Virus and Mouse Mammary Tumor Virus with Breast Cancer in Mexican Women. Sci. Rep. 2013;3:2970. doi: 10.1038/srep02970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tabriz H.M., Zendehdel K., Shahsiah R., Fereidooni F., Mehdipour B., Hosseini Z.M. Lack of Detection of the Mouse Mammary Tumor-like Virus (MMTV) Env Gene in Iranian Women Breast Cancer Using Real Time PCR. Asian Pac. J. Cancer Prev. 2013;14:2945–2948. doi: 10.7314/apjcp.2013.14.5.2945. [DOI] [PubMed] [Google Scholar]
- 111.Mazzanti C.M., Al Hamad M., Fanelli G., Scatena C., Zammarchi F., Zavaglia K., Lessi F., Pistello M., Naccarato A.G., Bevilacqua G. A Mouse Mammary Tumor Virus Env-like Exogenous Sequence Is Strictly Related to Progression of Human Sporadic Breast Carcinoma. Am. J. Pathol. 2011;179:2083–2090. doi: 10.1016/j.ajpath.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Park D.J., Southey M.C., Giles G.G., Hopper J.L. No Evidence of MMTV-like Env Sequences in Specimens from the Australian Breast Cancer Family Study. Breast Cancer Res. Treat. 2011;125:229–235. doi: 10.1007/s10549-010-0946-4. [DOI] [PubMed] [Google Scholar]
- 113.Lawson J.S., Glenn W.K., Salmons B., Ye Y., Heng B., Moody P., Johal H., Rawlinson W.D., Delprado W., Lutze-Mann L., et al. Mouse Mammary Tumor Virus–like Sequences in Human Breast Cancer. Cancer Res. 2010;70:3576–3585. doi: 10.1158/0008-5472.CAN-09-4160. [DOI] [PubMed] [Google Scholar]
- 114.Fukuoka H., Moriuchi M., Yano H., Nagayasu T., Moriuchi H. No Association of Mouse Mammary Tumor Virus-Related Retrovirus with Japanese Cases of Breast Cancer. J. Med. Virol. 2008;80:1447–1451. doi: 10.1002/jmv.21247. [DOI] [PubMed] [Google Scholar]
- 115.Hachana M., Trimeche M., Ziadi S., Amara K., Gaddas N., Mokni M., Korbi S. Prevalence and Characteristics of the MMTV-like Associated Breast Carcinomas in Tunisia. Cancer Lett. 2008;271:222–230. doi: 10.1016/j.canlet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 116.Cedro-Tanda A., Córdova-Solis A., Juárez-Cedillo T., Pina-Jiménez E., Hernández-Caballero M.E., Moctezuma-Meza C., Castelazo-Rico G., Gómez-Delgado A., Monsalvo-Reyes A.C., Salamanca-Gómez F.A., et al. Prevalence of HMTV in Breast Carcinomas and Unaffected Tissue from Mexican Women. BMC Cancer. 2014;14:942. doi: 10.1186/1471-2407-14-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Perzova R., Abbott L., Benz P., Landas S., Khan S., Glaser J., Cunningham C.K., Poiesz B. Is MMTV Associated with Human Breast Cancer? Maybe, but Probably Not. Virol. J. 2017;14:196. doi: 10.1186/s12985-017-0862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang Y., Holland J.F., Bleiweiss I.J., Melana S., Liu X., Pelisson I., Cantarella A., Stellrecht K., Mani S., Pogo B.G. Detection of Mammary Tumor Virus Env Gene-like Sequences in Human Breast Cancer. Cancer Res. 1995;55:5173–5179. [PubMed] [Google Scholar]
- 119.Hareuveni M., Lathe R. Breast Cancer Sequences Identified by Mouse Mammary Tumor Virus (MMTV) Antiserum Are Unrelated to MMTV. Int. J. Cancer. 1990;46:1134–1135. doi: 10.1002/ijc.2910460631. [DOI] [PubMed] [Google Scholar]
- 120.Mesa-Tejada R., Keydar I., Ramanarayanan M., Ohno T., Fenoglio C., Spiegelman S. Immunohistochemical Detection of a Cross-Reacting Virus Antigen in Mouse Mammary Tumors and Human Breast Carcinomas. J. Histochem. Cytochem. 1978;26:532–541. doi: 10.1177/26.7.80418. [DOI] [PubMed] [Google Scholar]
- 121.Kordon E.C. MMTV-Induced Pregnancy-Dependent Mammary Tumors: Early History and New Perspectives. J. Mammary Gland. Biol. Neoplasia. 2008;13:289–297. doi: 10.1007/s10911-008-9091-7. [DOI] [PubMed] [Google Scholar]
- 122.Pogo B.G.-T., Holland J.F., Levine P.H. Human Mammary Tumor Virus in Inflammatory Breast Cancer. Cancer. 2010;116:2741–2744. doi: 10.1002/cncr.25179. [DOI] [PubMed] [Google Scholar]
- 123.Khalid H.F., Bibi S., Ali A., Fawad N., Shams M.U., Idrees W., Waqas M., Rafique S., Khan A., Khan F., et al. Decoding the Mystery of MMTV-like Virus and Its Relationship with Breast Cancer Metastasis. J. Infect. Public Health. 2023;16:1396–1402. doi: 10.1016/j.jiph.2023.07.004. [DOI] [PubMed] [Google Scholar]
- 124.Ye Z., Chen L., Zhong H., Cao L., Fu P., Xu J. Epidemiology and Clinical Characteristics of Epstein-Barr Virus Infection among Children in Shanghai, China, 2017–2022. Front. Cell. Infect. Microbiol. 2023;13:1139068. doi: 10.3389/fcimb.2023.1139068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smatti M.K., Al-Sadeq D.W., Ali N.H., Pintus G., Abou-Saleh H., Nasrallah G.K. Epstein–Barr Virus Epidemiology, Serology, and Genetic Variability of LMP-1 Oncogene Among Healthy Population: An Update. Front. Oncol. 2018;8:211. doi: 10.3389/fonc.2018.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lin J.-H., Tsai C.-H., Chu J.-S., Chen J.-Y., Takada K., Shew J.-Y. Dysregulation of HER2/HER3 Signaling Axis in Epstein-Barr Virus-Infected Breast Carcinoma Cells. J. Virol. 2007;81:5705–5713. doi: 10.1128/JVI.00076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lorenzetti M.A., De Matteo E., Gass H., Martinez Vazquez P., Lara J., Gonzalez P., Preciado M.V., Chabay P.A. Characterization of Epstein Barr Virus Latency Pattern in Argentine Breast Carcinoma. PLoS ONE. 2010;5:e13603. doi: 10.1371/journal.pone.0013603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gouadfel K., Khenchouche A., Rabea S., Mansour A.A., Salem-Bekhit M.M., Ouhida S., Msela A., Salem M.M., Erto A., Benguerba Y., et al. The Potential Role of Epstein-Barr Virus in Breast Cancer Development. Cell. Mol. Biol. 2023;69:241–249. doi: 10.14715/cmb/2023.69.13.36. [DOI] [PubMed] [Google Scholar]
- 129.Mekrazi S., Kallel I., Jamai D., Yengui M., Khabir A., Gdoura R. Epstein-Barr Virus in Breast Carcinoma and in Triple Negative Cases Impact on Clinical Outcomes. Pathol. Res. Pract. 2023;245:154484. doi: 10.1016/j.prp.2023.154484. [DOI] [PubMed] [Google Scholar]
- 130.Shahi V., Agarwal P., Qayoom S., Kumar V., Tewari S., Raghuvanshi S., Singh U.S., Goel M.M. Detection of Epstein Barr Nuclear Antigen-1 (EBNA-1), Early Antigen 1F, 2R (EA-1F, EA- 2R) along with Epstein-Barr Virus Latent Membrane Protein 1 (LMP1) in Breast Cancer of Northern India: An Interim Analysis. Asian Pac. J. Cancer Prev. 2022;23:3717–3723. doi: 10.31557/APJCP.2022.23.11.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang W.-T., Zhu G.-L., Xu W.-Q., Zhang W., Wang H.-Z., Wang Y.-B., Li Y.-X. Association of PD-1/PD-L1 Expression and Epstein--Barr Virus Infection in Patients with Invasive Breast Cancer. Diagn. Pathol. 2022;17:61. doi: 10.1186/s13000-022-01234-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ghaffari H., Tavakoli A., Nafissi N., Farahmand M., Ghorbani S., Moochani S.S., Hashemi-Bahremani M., Alebouyeh M.R., Monavari S.H. Human Cytomegalovirus and Epstein-Barr Virus Infections in Breast Cancer: A Molecular Study on Iranian Women. Breast Dis. 2021;40:227–233. doi: 10.3233/BD-201019. [DOI] [PubMed] [Google Scholar]
- 133.Charostad J., Nakhaei M., Azaran A., Kaydani G.A., Astani A., Motamedfar A., Makvandi M. MiRNA-218 Is Frequently Downregulated in Malignant Breast Tumors: A Footprint of Epstein-Barr Virus Infection. Iran. J. Pathol. 2021;16:376–385. doi: 10.30699/IJP.20201.521107.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Golrokh Mofrad M., Kazeminezhad B., Faghihloo E. Prevalence of Epstein-Barr Virus (EBV) in Iranian Breast Carcinoma Patients. Asian Pac. J. Cancer Prev. 2020;21:133–137. doi: 10.31557/APJCP.2020.21.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mostafaei S., Vahidi Manesh P., Sadri Nahand J., Nesaei A., Sorayyayi S., Abasabadi F., Mirzaei H., Etemadi A., O’ Neill A., Donnelly S.C., et al. The Role of Epstein-Barr Virus-Expressed Genes in Breast Cancer Development. Breast J. 2020;26:2323–2326. doi: 10.1111/tbj.14021. [DOI] [PubMed] [Google Scholar]
- 136.Dowran R., Joharinia N., Safaei A., Bakhtiyarizadeh S., Alidadi Soleimani A., Alizadeh R., Mir-Shiri S., Sarvari J. No Detection of EBV, BKV and JCV in Breast Cancer Tissue Samples in Iran. BMC Res. Notes. 2019;12:171. doi: 10.1186/s13104-019-4178-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sharifpour C., Makvandi M., Samarbafzadeh A., Talaei-Zadeh A., Ranjbari N., Nisi N., Azaran A., Jalilian S., Varnaseri M., Pirmoradi R., et al. Frequency of Epstein–Barr Virus DNA in Formalin-Fixed Paraffin-Embedded Tissue of Patients with Ductal Breast Carcinoma. Asian Pac. J. Cancer Prev. 2019;20:687–692. doi: 10.31557/APJCP.2019.20.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pai T., Gupta S., Gurav M., Nag S., Shet T., Patil A., Desai S. Evidence for the Association of Epstein-Barr Virus in Breast Cancer in Indian Patients Using in-Situ Hybridization Technique. Breast J. 2018;24:16–22. doi: 10.1111/tbj.12828. [DOI] [PubMed] [Google Scholar]
- 139.El-Naby N.E.H., Hassan Mohamed H., Mohamed Goda A., El Sayed Mohamed A. Epstein-Barr Virus Infection and Breast Invasive Ductal Carcinoma in Egyptian Women: A Single Center Experience. J. Egypt. Natl. Cancer Inst. 2017;29:77–82. doi: 10.1016/j.jnci.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 140.Aboulkassim T., Yasmeen A., Akil N., Batist G., Al Moustafa A.-E. Incidence of Epstein-Barr Virus in Syrian Women with Breast Cancer: A Tissue Microarray Study. Hum. Vaccin. Immunother. 2015;11:951–955. doi: 10.1080/21645515.2015.1009342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ballard A.J. Epstein-Barr Virus Infection Is Equally Distributed across the Invasive Ductal and Invasive Lobular Forms of Breast Cancer. Pathol. Res. Pract. 2015;211:1003–1005. doi: 10.1016/j.prp.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 142.Richardson A.K., Currie M.J., Robinson B.A., Morrin H., Phung Y., Pearson J.F., Anderson T.P., Potter J.D., Walker L.C. Cytomegalovirus and Epstein-Barr Virus in Breast Cancer. PLoS ONE. 2015;10:e0118989. doi: 10.1371/journal.pone.0118989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Marrão G., Habib M., Paiva A., Bicout D., Fallecker C., Franco S., Fafi-Kremer S., Simões da Silva T., Morand P., Freire de Oliveira C., et al. Epstein-Barr Virus Infection and Clinical Outcome in Breast Cancer Patients Correlate with Immune Cell TNF-α/IFN-γ Response. BMC Cancer. 2014;14:665. doi: 10.1186/1471-2407-14-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baltzell K., Buehring G.C., Krishnamurthy S., Kuerer H., Shen H.M., Sison J.D. Epstein-Barr Virus Is Seldom Found in Mammary Epithelium of Breast Cancer Tissue Using in Situ Molecular Methods. Breast Cancer Res. Treat. 2012;132:267–274. doi: 10.1007/s10549-011-1841-3. [DOI] [PubMed] [Google Scholar]
- 145.Zekri A.-R.N., Bahnassy A.A., Mohamed W.S., El-Kassem F.A., El-Khalidi S.J., Hafez M.M., Hassan Z.K. Epstein-Barr Virus and Breast Cancer: Epidemiological and Molecular Study on Egyptian and Iraqi Women. J. Egypt. Natl. Cancer Inst. 2012;24:123–131. doi: 10.1016/j.jnci.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 146.Hachana M., Amara K., Ziadi S., Romdhane E., Gacem R.B., Trimeche M. Investigation of Epstein-Barr Virus in Breast Carcinomas in Tunisia. Pathol. Res. Pract. 2011;207:695–700. doi: 10.1016/j.prp.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 147.Kadivar M., Monabati A., Joulaee A., Hosseini N. Epstein-Barr Virus and Breast Cancer: Lack of Evidence for an Association in Iranian Women. Pathol. Oncol. Res. 2011;17:489–492. doi: 10.1007/s12253-010-9325-z. [DOI] [PubMed] [Google Scholar]
- 148.Mazouni C., Fina F., Romain S., Ouafik L., Bonnier P., Brandone J.-M., Martin P.-M. Epstein-Barr Virus as a Marker of Biological Aggressiveness in Breast Cancer. Br. J. Cancer. 2011;104:332–337. doi: 10.1038/sj.bjc.6606048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Joshi D., Quadri M., Gangane N., Joshi R., Gangane N. Association of Epstein Barr Virus Infection (EBV) with Breast Cancer in Rural Indian Women. PLoS ONE. 2009;4:e8180. doi: 10.1371/journal.pone.0008180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fawzy S., Sallam M., Awad N.M. Detection of Epstein-Barr Virus in Breast Carcinoma in Egyptian Women. Clin. Biochem. 2008;41:486–492. doi: 10.1016/j.clinbiochem.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 151.Khan G., Philip P.S., Al Ashari M., Houcinat Y., Daoud S. Localization of Epstein-Barr Virus to Infiltrating Lymphocytes in Breast Carcinomas and Not Malignant Cells. Exp. Mol. Pathol. 2011;91:466–470. doi: 10.1016/j.yexmp.2011.04.018. [DOI] [PubMed] [Google Scholar]
- 152.Glaser S.L., Canchola A.J., Keegan T.H.M., Clarke C.A., Longacre T.A., Gulley M.L. Variation in Risk and Outcomes of Epstein-Barr Virus-Associated Breast Cancer by Epidemiologic Characteristics and Virus Detection Strategies: An Exploratory Study. Cancer Causes Control. 2017;28:273–287. doi: 10.1007/s10552-017-0865-3. [DOI] [PubMed] [Google Scholar]
- 153.Mashaly M., Ghorab D., Hegazy M., Abdelkhalek M., Gaballah K., Elzehery R. Association between Epstein-Barr Virus Gene Polymorphism and Breast Cancer Risk among Egyptian Females. Asian Pac. J. Cancer Prev. 2022;23:641–650. doi: 10.31557/APJCP.2022.23.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Heng Y.J., Love S., DeHart J.C., Fingeroth J.D., Wulf G.M. The Association of Infectious Mononucleosis and Invasive Breast Cancer in The Health of Women (HOW) Study®. Breast Cancer. 2022;29:731–739. doi: 10.1007/s12282-022-01351-3. [DOI] [PubMed] [Google Scholar]
- 155.Yang T., Liu D., Fang S., Ma W., Wang Y. Cytomegalovirus and Glioblastoma: A Review of the Biological Associations and Therapeutic Strategies. J. Clin. Med. 2022;11:5221. doi: 10.3390/jcm11175221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Herbein G. Tumors and Cytomegalovirus: An Intimate Interplay. Viruses. 2022;14:812. doi: 10.3390/v14040812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kumar A., Tripathy M.K., Pasquereau S., Al Moussawi F., Abbas W., Coquard L., Khan K.A., Russo L., Algros M.-P., Valmary-Degano S., et al. The Human Cytomegalovirus Strain DB Activates Oncogenic Pathways in Mammary Epithelial Cells. eBioMedicine. 2018;30:167–183. doi: 10.1016/j.ebiom.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Herbein G. High-Risk Oncogenic Human Cytomegalovirus. Viruses. 2022;14:2462. doi: 10.3390/v14112462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bouezzedine F., El Baba R., Haidar Ahmad S., Herbein G. Polyploid Giant Cancer Cells Generated from Human Cytomegalovirus-Infected Prostate Epithelial Cells. Cancers. 2023;15:4994. doi: 10.3390/cancers15204994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nehme Z., Pasquereau S., Haidar Ahmad S., El Baba R., Herbein G. Polyploid Giant Cancer Cells, EZH2 and Myc Upregulation in Mammary Epithelial Cells Infected with High-Risk Human Cytomegalovirus. EBioMedicine. 2022;80:104056. doi: 10.1016/j.ebiom.2022.104056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.El Baba R., Pasquereau S., Haidar Ahmad S., Diab-Assaf M., Herbein G. Oncogenic and Stemness Signatures of the High-Risk HCMV Strains in Breast Cancer Progression. Cancers. 2022;14:4271. doi: 10.3390/cancers14174271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Touma J., Pantalone M.R., Rahbar A., Liu Y., Vetvik K., Sauer T., Söderberg-Naucler C., Geisler J. Human Cytomegalovirus Protein Expression Is Correlated with Shorter Overall Survival in Breast Cancer Patients: A Cohort Study. Viruses. 2023;15:732. doi: 10.3390/v15030732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nakhaie M., Charostad J., Azaran A., Arabzadeh S.A.M., Motamedfar A., Iranparast S., Ahmadpour F., Talaeizadeh A., Makvandi M. Molecular and Serological Prevalence of HCMV in Iranian Patients with Breast Cancer. Asian Pac. J. Cancer Prev. 2021;22:2011–2016. doi: 10.31557/APJCP.2021.22.7.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Touma J., Liu Y., Rahbar A., Pantalone M.R., Almazan N.M., Vetvik K., Söderberg-Nauclér C., Geisler J., Sauer T. Detection of Human Cytomegalovirus Proteins in Paraffin-Embedded Breast Cancer Tissue Specimens—A Novel, Automated Immunohistochemical Staining Protocol. Microorganisms. 2021;9:1059. doi: 10.3390/microorganisms9051059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Costa H., Touma J., Davoudi B., Benard M., Sauer T., Geisler J., Vetvik K., Rahbar A., Söderberg-Naucler C. Human Cytomegalovirus Infection Is Correlated with Enhanced Cyclooxygenase-2 and 5-Lipoxygenase Protein Expression in Breast Cancer. J. Cancer Res. Clin. Oncol. 2019;145:2083–2095. doi: 10.1007/s00432-019-02946-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sepahvand P., Makvandi M., Samarbafzadeh A., Talaei-Zadeh A., Ranjbari N., Nisi N., Azaran A., Jalilian S., Pirmoradi R., Makvandi K., et al. Human Cytomegalovirus DNA among Women with Breast Cancer. Asian Pac. J. Cancer Prev. 2019;20:2275–2279. doi: 10.31557/APJCP.2019.20.8.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.El Shazly D.F., Bahnassey A.A., Omar O.S., Elsayed E.T., Al-Hindawi A., El-Desouky E., Youssef H., Zekri A.-R.N. Detection of Human Cytomegalovirus in Malignant and Benign Breast Tumors in Egyptian Women. Clin. Breast Cancer. 2018;18:e629–e642. doi: 10.1016/j.clbc.2017.10.018. [DOI] [PubMed] [Google Scholar]
- 168.El-Shinawi M., Mohamed H.T., El-Ghonaimy E.A., Tantawy M., Younis A., Schneider R.J., Mohamed M.M. Human Cytomegalovirus Infection Enhances NF-κB/P65 Signaling in Inflammatory Breast Cancer Patients. PLoS ONE. 2013;8:e55755. doi: 10.1371/journal.pone.0055755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pandey J.P., Gao G., Namboodiri A.M., Iwasaki M., Kasuga Y., Hamada G.S., Tsugane S. Humoral Immunity to Cytomegalovirus Glycoprotein B in Patients with Breast Cancer and Matched Controls: Contribution of Immunoglobulin γ, κ, and Fcγ Receptor Genes. J. Infect. Dis. 2016;213:611–617. doi: 10.1093/infdis/jiv472. [DOI] [PubMed] [Google Scholar]
- 170.Mendoza W., Isaza J.P., López L., López-Herrera A., Gutiérrez L.A. Bovine Leukemia Virus Detection in Humans: A Systematic Review and Meta-Analysis. Virus Res. 2023;335:199186. doi: 10.1016/j.virusres.2023.199186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Amato S., Ramsey J., Ahern T.P., Rovnak J., Barlow J., Weaver D., Eyasu L., Singh R., Cintolo-Gonzalez J. Exploring the Presence of Bovine Leukemia Virus among Breast Cancer Tumors in a Rural State. Breast Cancer Res. Treat. 2023;202:325–334. doi: 10.1007/s10549-023-07061-4. [DOI] [PubMed] [Google Scholar]
- 172.Yamanaka M.P., Saito S., Hara Y., Matsuura R., Takeshima S.-N., Hosomichi K., Matsumoto Y., Furuta R.A., Takei M., Aida Y. No Evidence of Bovine Leukemia Virus Proviral DNA and Antibodies in Human Specimens from Japan. Retrovirology. 2022;19:7. doi: 10.1186/s12977-022-00592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Adekanmbi F., McNeely I., Omeler S., Kalalah A., Poudel A., Merner N., Wang C. Absence of Bovine Leukemia Virus in the Buffy Coats of Breast Cancer Cases from Alabama, USA. Microb. Pathog. 2021;161:105238. doi: 10.1016/j.micpath.2021.105238. [DOI] [PubMed] [Google Scholar]
- 174.Olaya-Galán N.N., Salas-Cárdenas S.P., Rodriguez-Sarmiento J.L., Ibáñez-Pinilla M., Monroy R., Corredor-Figueroa A.P., Rubiano W., de la Peña J., Shen H., Buehring G.C., et al. Risk Factor for Breast Cancer Development under Exposure to Bovine Leukemia Virus in Colombian Women: A Case-Control Study. PLoS ONE. 2021;16:e0257492. doi: 10.1371/journal.pone.0257492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Delarmelina E., Buzelin M.A., de Souza B.S., Souto F.M., Bicalho J.M., Câmara R.J.F., Resende C.F., Bueno B.L., Victor R.M., Galinari G.C.F., et al. High Positivity Values for Bovine Leukemia Virus in Human Breast Cancer Cases from Minas Gerais, Brazil. PLoS ONE. 2020;15:e0239745. doi: 10.1371/journal.pone.0239745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Schwingel D., Andreolla A.P., Erpen L.M.S., Frandoloso R., Kreutz L.C. Bovine Leukemia Virus DNA Associated with Breast Cancer in Women from South Brazil. Sci. Rep. 2019;9:2949. doi: 10.1038/s41598-019-39834-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Buehring G.C., Shen H., Schwartz D.A., Lawson J.S. Bovine Leukemia Virus Linked to Breast Cancer in Australian Women and Identified before Breast Cancer Development. PLoS ONE. 2017;12:e0179367. doi: 10.1371/journal.pone.0179367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gillet N.A., Willems L. Whole Genome Sequencing of 51 Breast Cancers Reveals that Tumors Are Devoid of Bovine Leukemia Virus DNA. Retrovirology. 2016;13:75. doi: 10.1186/s12977-016-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Zhang R., Jiang J., Sun W., Zhang J., Huang K., Gu X., Yang Y., Xu X., Shi Y., Wang C. Lack of Association between Bovine Leukemia Virus and Breast Cancer in Chinese Patients. Breast Cancer Res. 2016;18:101. doi: 10.1186/s13058-016-0763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Buehring G.C., Shen H.M., Jensen H.M., Jin D.L., Hudes M., Block G. Exposure to Bovine Leukemia Virus Is Associated with Breast Cancer: A Case-Control Study. PLoS ONE. 2015;10:e0134304. doi: 10.1371/journal.pone.0134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Buehring G.C., Shen H.M., Jensen H.M., Choi K.Y., Sun D., Nuovo G. Bovine Leukemia Virus DNA in Human Breast Tissue. Emerg. Infect. Dis. 2014;20:772–782. doi: 10.3201/eid2005.131298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Khan Z., Abubakar M., Arshed M.J., Aslam R., Sattar S., Shah N.A., Javed S., Tariq A., Bostan N., Manzoor S. Molecular Investigation of Possible Relationships Concerning Bovine Leukemia Virus and Breast Cancer. Sci. Rep. 2022;12:4161. doi: 10.1038/s41598-022-08181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Malekpour Afshar R., Mollaei H., Zandi B., Iranpour M. Evaluation of JC and Cytomegalo Viruses in Glioblastoma Tissue. Asian Pac. J. Cancer Prev. 2016;17:4907–4911. doi: 10.22034/APJCP.2016.17.11.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zheng H.-C., E Y., Cui Z.-G., Zhao S., Zhang Y. The Oncogenic Roles of JC Virus T Antigen in Breast Carcinogenesis. Front. Mol. Biosci. 2021;8:687444. doi: 10.3389/fmolb.2021.687444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Theodoropoulos G., Panoussopoulos D., Papaconstantinou I., Gazouli M., Perdiki M., Bramis J., Lazaris A.C. Assessment of JC Polyoma Virus in Colon Neoplasms. Dis. Colon Rectum. 2005;48:86–91. doi: 10.1007/s10350-004-0737-2. [DOI] [PubMed] [Google Scholar]
- 186.Murai Y., Zheng H., Aziz H.O.A., Mei H., Kutsuna T., Nakanishi Y., Tsuneyama K., Takano Y. High JC Virus Load in Gastric Cancer and Adjacent Non-cancerous Mucosa. Cancer Sci. 2007;98:25–31. doi: 10.1111/j.1349-7006.2006.00354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hachana M., Amara K., Ziadi S., Gacem R.B., Korbi S., Trimeche M. Investigation of Human JC and BK Polyomaviruses in Breast Carcinomas. Breast Cancer Res. Treat. 2012;133:969–977. doi: 10.1007/s10549-011-1876-5. [DOI] [PubMed] [Google Scholar]
- 188.Silling S., Kreuter A., Gambichler T., Meyer T., Stockfleth E., Wieland U. Epidemiology of Merkel Cell Polyomavirus Infection and Merkel Cell Carcinoma. Cancers. 2022;14:6176. doi: 10.3390/cancers14246176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Khan G., Philip P.S., Naase M., Al Zarouni K.M.I. No Evidence for the Involvement of XMRV or MCV in the Pathogenesis of Breast Cancer. Br. J. Cancer. 2012;106:1166–1170. doi: 10.1038/bjc.2012.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author.