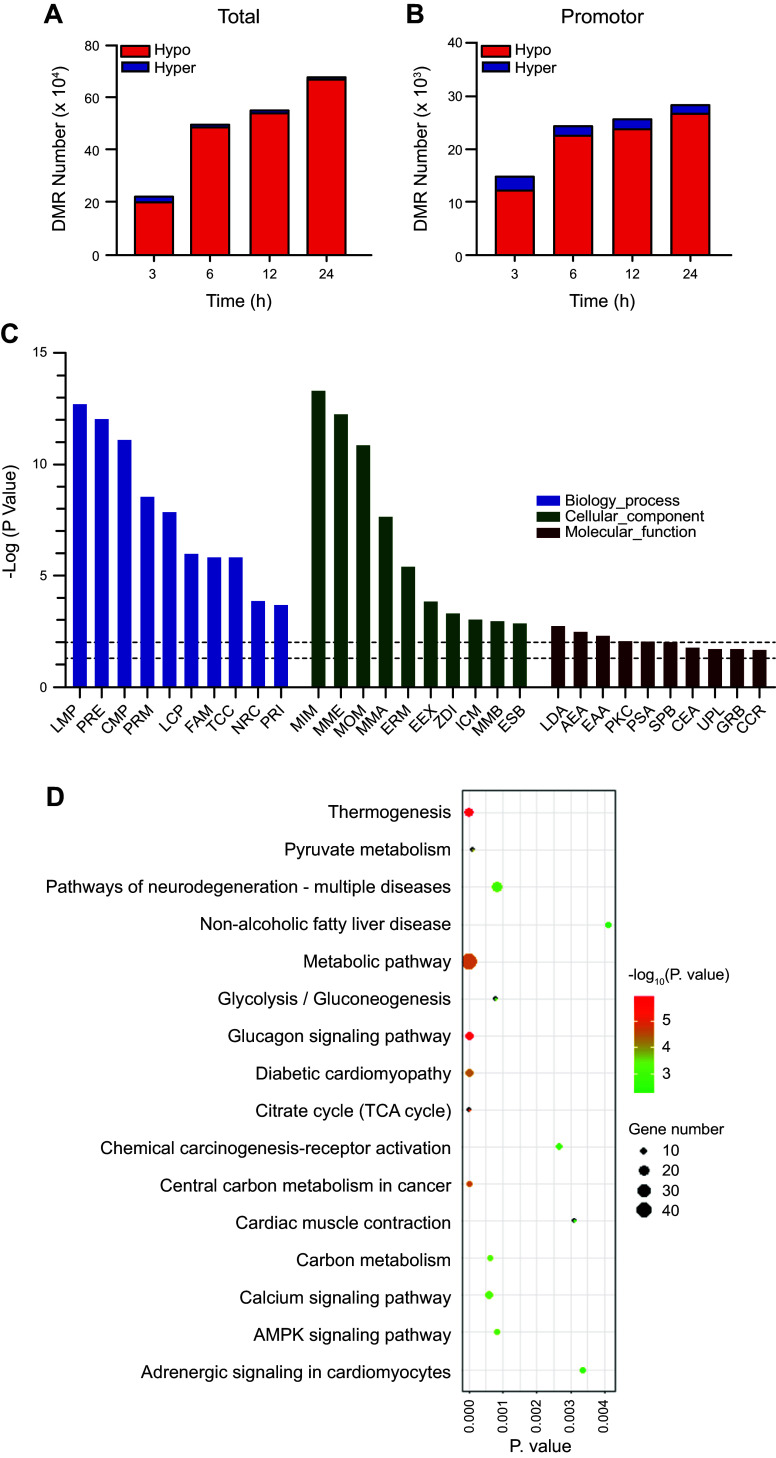
Figure 2.
Effects of cholestenoic acid (CA) on DNA methylation in hepatocytes using whole genome bisulfite sequencing (WGBS) analysis. HepG-2 cells were cultured in high glucose (HG) medium for 72 h and followed by treatment with 20 µM CA treatment for 0, 3, 6, 12, and 24 h. One microgram of genomic DNA was used to prepare libraries. A: number of differential methylated regions (DMRs) in whole genome. B: number of DMRs in promoter regions. C: top terms (P < 0.05) of Gene Ontology (GO) analysis, enriched in hypomethylated DMRs in promoter regions. LMP, lipid metabolic process; PRE, positive regulation of ERK1 and ERK2 cascade; CMP, carbohydrate metabolic process; PRM, positive regulation of MAPK cascade; LCP, lipid catabolic process; FAM, fatty acid metabolic process; TCC, tricarboxylic acid cycle; NRC, negative regulation of cell growth; PRI, positive regulation of interleukin-6 production; MIM, mitochondrial inner membrane; MME, mitochondrial membrane; MOM, mitochondrial outer membrane; MMA, mitochondrial matrix; ERM, endoplasmic reticulum membrane; EEX, extracellular exosome; ADI, Z disc; ICM, integral component of mitochondrial inner membrane; MMB, membrane; ESP, extracellular space; LDA, l-lactate dehydrogenase activity; AEA, 1-alkyl-2-acetylglycerophosphocholine esterase activity; EAA, enzyme activator activity; PKC, protein kinase A catalytic subunit binding; PSA, protein self-association; SPB, S100 protein binding; CEA, cysteine-type endopeptidase activator activity involved in apoptotic process; UPL, ubiquitin protein ligase binding; GRB, GABA receptor binding; CCR: complement component C5a receptor activity. D: top significantly (P < 0.05) enriched Kyoto Encyclopedia of Genes and Genome (KEGG) pathways of promoter region with hypomethylated DMRs.