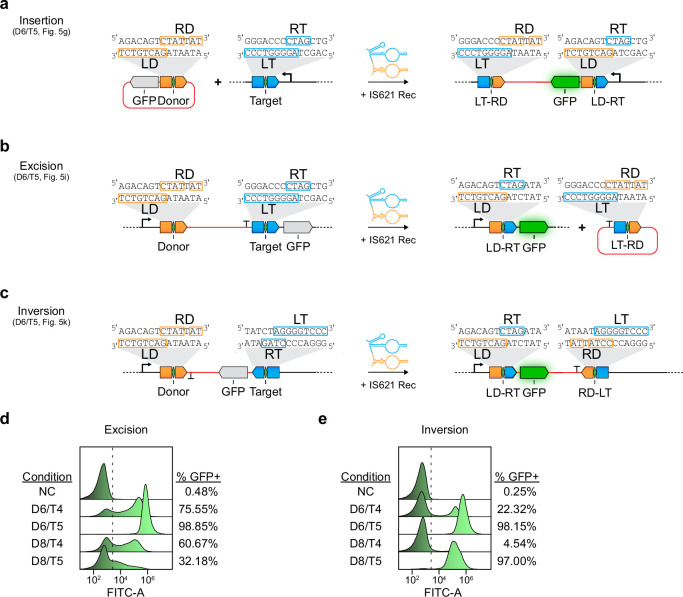
Extended Data Fig. 9. Insertion, excision and inversion using the IS621 bridge recombination system.
a, Schematic of the insertion reaction. Insertion takes place when the target and donor sequences are on different DNA molecules. The orientation of the insertion can be controlled by the strand placement of the target and the donor. b, Schematic of the excision reaction. Excision can occur when the target and donor sequences exist on the same molecule and in the same orientation (i.e. LD and LT are on the same strand). c, Schematic of the inversion reaction. Inversion can be catalysed when the target and donor sequences exist on the same molecule, but in the opposing orientation (i.e. on opposite strands). d, DNA excision in E. coli with reprogrammed bridge RNAs. The distribution of FITC-A signal for the cell population is shown, with a representative gating strategy for evaluating the percentage of GFP+ cells. The donor-target pair is given. Negative control (NC) expresses the reporter with no target or donor, the recombinase, and no bridge RNA. Plots are representative of 3 replicates featured in Fig. 5j. e, DNA inversion in E. coli with reprogrammed bridge RNAs. The distribution of FITC-A signal for the cell population is shown, with a representative gating strategy for evaluating the percentage of GFP+ cells. The donor-target pair is given. Negative control (NC) expresses the reporter with no target or donor, the recombinase, and no bridge RNA. Plots are representative of 3 replicates featured in Fig. 5k.