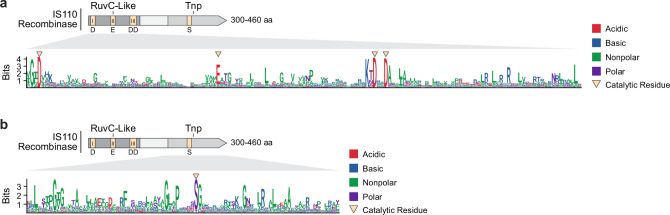
Extended Data Fig. 1. Conserved residues in the RuvC-like and Tnp domains of IS110.
a, Sequence logo of 213,171 aligned RuvC-like domains identified in IS110 protein sequences. The RuvC-like and Tnp domains shown here were identified using hmmsearch and Pfam models DEDD_Tnp_IS110 (PF01548.19) and Transposase_20 (PF02371.18), respectively. RuvC-like domains were aligned using hmmalign, and these alignments were visualized to identify conserved residues. The conserved residues of the characteristic DEDD motif are highlighted with an arrowhead. The y-axis indicates entropy at each position as measured in bits, with log220 ≈ 4.32 bits being maximally conserved. b, Sequence logo of 208,634 aligned Tnp domains identified in IS110 protein sequences. The Tnp domains were identified, extracted, and analysed using the same procedure as for the RuvC-like domain. A highly conserved serine residue is highlighted with an arrowhead. The y-axis indicates entropy at each position as measured in bits, with log220 ≈ 4.32 bits being maximally conserved.