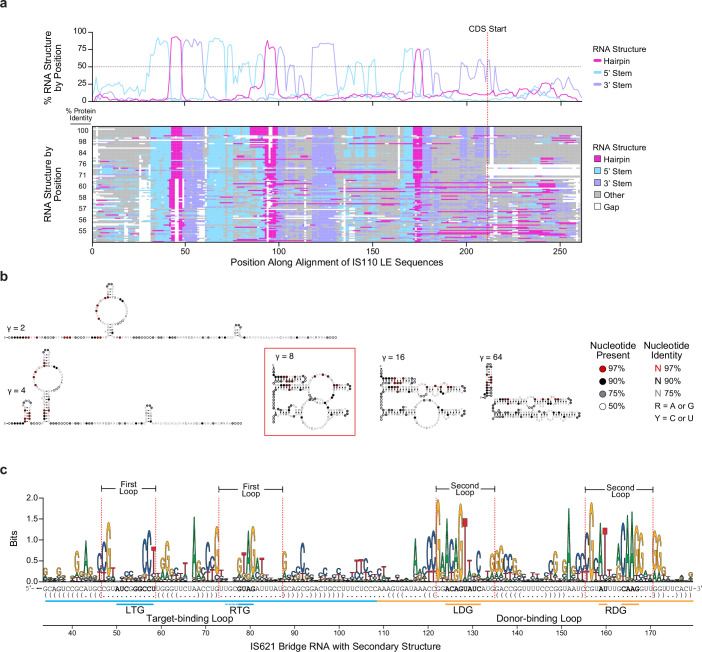
Extended Data Fig. 3. Secondary structure alignment of IS621 non-coding ends and consensus secondary structure prediction.
a, Secondary RNA structure alignment of the LE of 103 orthologues of IS621. Secondary RNA structures of the LE of 103 orthologues are predicted and aligned by cluster identity. The percentage of each position corresponding to a 5′ stem, hairpin, or 3′ stem are plotted with a dotted line indicating structures that are conserved in over 50% of sequences. For LE sequences shown along the y-axis, the similarity of their cognate proteins relative to the IS621 recombinase is indicated. This type of visualization was often used throughout the study to determine the presence or absence of a structured ncRNA sequence in the flanks of IS110 recombinase ORFs. b, RNA structures predicted from the LE sequence alignment in a. RNA structures were predicted using ConsAliFold, which uses a parameter γ to control the prediction balance between positive values (or sequence alignment column base-pairings) and negative values (or unpaired sequence alignment columns). Higher values of γ result in more predicted base-pairing. Showing structures resulting from γ = 2, γ = 4, γ = 8, γ = 16, and γ = 64. The value γ = 8 was used for the initial IS621 ncRNA model in this study. c, Nucleotide conservation across the predicted ncRNA. 2,715 ncRNA orthologue sequences were identified using an iterative search with the original IS621 model, and then aligned with cmalign. The x-axis indicates conservation of nucleotides as measured in bits, quantifying entropy. Highlighting the regions within the prominent internal loops with dotted red lines, with dot-bracket RNA secondary structure notation along the x-axis. The first loop has low sequence conservation (average information content = 0.48 ± 0.09), while the second one is much more conserved (average information content = 0.93 ± 0.11). Sequence features of the bridge RNA are highlighted for clarity.