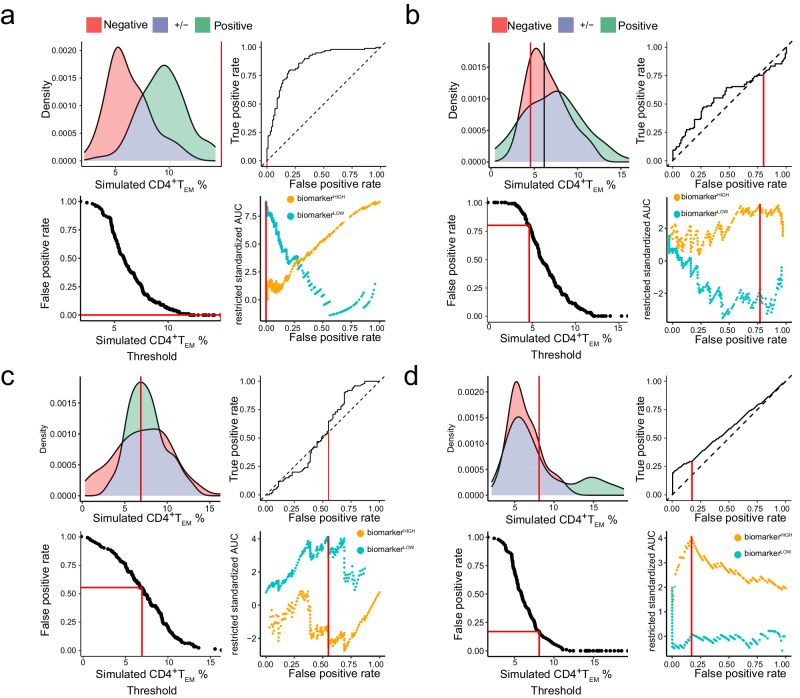
Fig. 5. Restriction of synthesised flow cytometry datasets.
We present examples of biomarker distributions in two classes, which are intended to represent sets of patients with different clinical outcomes. The distribution of values from positive (i.e. diseased) class are coloured green and values from negative (i.e. control) class are coloured red; overlapping density areas are coloured purple. For each example, we present the following: a plot of positive and negative class densities; the complete ROC curve; a plot of biomarker values against FPR; a plot of rzAUC calculated for biomarkerHIGH (orange) and biomarkerLOW (blue) samples at all FPR values. In each plot, red lines indicate the optimal restriction as a biomarker value or FPR value. a A synthetic example of a symmetrical ROC curve from 100 negative and 100 positive samples. b A synthetic example of a right-skewed ROC curve from 100 negative and 100 positive samples. c A synthetic example of a left-skewed ROC curve from 100 negative and 100 positive samples. d Results for a synthetic right-skewed ROC curve from 100 negative samples and 100 positive samples from a bimodally distributed positive population. In this example, the positive population comprises 20% cases with elevated biomarker expression and 80% cases with unaltered biomarker expression .